Manuscript accepted on :18-11-2021
Published online on: 22-11-2021
Plagiarism Check: Yes
Reviewed by: Dr. Sabu Thomas

Second Review by: Dr. Salman Ahmed Pharmacognosy

Final Approval by: Dr. Ayush Dogra
Charlotte Jessica Fernandes , Bhavya Doddavarapu
, Bhavya Doddavarapu , Anupama Harry
, Anupama Harry , Sri Priya Srikakulam Dilip
, Sri Priya Srikakulam Dilip and Lokesh Ravi*
and Lokesh Ravi*
Department of Botany, St. Joseph’s College (Autonomous), Bengaluru-27 India
Corresponding Author E-mail: lokesh.ravi@sjc.ac.in
DOI : https://dx.doi.org/10.13005/bpj/2326
Abstract
Given the rising demand for biological pigments, especially of microbial origin – the present study was conducted so as to report a potential source for the extraction of microbial pigment. The main objective was to isolate and identify a pigment–producing actinomycete because pigment production is prevelant in this group. A powdery, greenish–blue colony with a chalky azure aerial mass was isolated from one of the many rhizosphere soil samples. Upon preliminary investigation, viz. colony characterization and grams staining, the suspected colony was observed to have a filamentous margin with a slightly raised elevation and gram–positive filamentous hyphae.Biochemical analyses of the organism revealed positive results for carbohydrate fermentation and Triple Sugar Iron (TSI) test with no signs of gas production during the former but gas & H2S production during the latter. The identity of the isolate was established via 16S rDNA and phylogeny analysis, which strongly suggested it was Saccharomonospora azurea. Limited research pertaining to morphology, physiology, genomics and secondary metabolite production with no reports on the physicochemical properties of the pigment produced by S. azureaadequately suggests that it is relatively novel. Hence, further studies related to the same could be beneficial to the scientific community.
Keywords
Actinobacteria; actinomycetes; Greenish-Blue Pigment; Microbial Pigment; Rhizosphere Soil; Saccharomonospora azurea
Download this article as:| Copy the following to cite this article: Fernandes C. J, Doddavarapu B, Harry A, Dilip S. P. S, Ravi L. Isolation and Identification of Pigment Producing Actinomycete Saccharomonospora azurea SJCJABS01. Biomed Pharmacol J 2021;14(4). |
| Copy the following to cite this URL: Fernandes C. J, Doddavarapu B, Harry A, Dilip S. P. S, Ravi L. Isolation and Identification of Pigment Producing Actinomycete Saccharomonospora azurea SJCJABS01. Biomed Pharmacol J 2021;14(4). Available from: https://bit.ly/3CK7fCp |
Introduction
Actinobacteria are a ubiquitous, heterogenous group of Gram–positive bacteria characterized by their fungal morphology and high GC content (>55 mol %)1–3. These bacteria exhibit physiological diversity as apparent from their ability to produce, synthesize, and excrete numerous primary and secondary metabolites1. As a result of their extensive physiological diversity, actinomycetes have great biotechnological potential. One such note worthy metabolite originating from actinomycetes are pigments. Actinomycetes are capable of synthesizing a wide range of dark pigments referred to as melanin/melanoid pigments4, some well-established pigments from actinomycetes are-blue (Streptomyces coelicolor)5, brown (Streptomyces sp.)6, green (Actinomyces viscosus and Saccharomonospora viridis)2,7, orange (A.naeslundii)2, red (Streptomyces echinoruber)8, 9, violet (Streptomyces mauvecolor)10, yellow (Streptomyces hygroscopicus)11, etc. Pigment production is influenced by chemical, physical and physiological factors3 such as anaerobic conditions and low temperatures (28 – 30°C), that boost pigment production in Streptomyces coelicolor5. Pigments are considered to be a helpful criterion for taxonomical investigations. Testing melanin production by using L-tyrosine/L-DOPA as a substrate is considered to be a potential criterion for the identification and classification of Streptomyces4. Besides their significance in taxonomic studies, these microbial pigments have a broad spectrum of biological activities such as antibiotic, antimicrobial, antioxidant, and antitumor12. With rising demands for biological pigments over synthetic colorants, there is a lot of work tending towards the identification and utilization of pigments and the microorganism producing them. Since pigment production seems to be more widely present in actinomycetes than in any other microbial groups, they are considered as the quintessential targets13.
The genus Saccharomonospora comprises of a group of bacteria that are of keen interest as their genome is sofar poorly characterized. The bacteria of this genus occur in diverse habitats (compost, leaf litter, manure, peat, etc.,) and are assumed to play a role in the primary degradation of plant material by digesting hemicellulose14. Nonomura & Ohara (1971) described the genus Saccharomonospora for bacteria that predominantly produced single spores, and occasionally spores in pairs and short chains, on aerial hyphae. Members of this genus contain a type IV cell wall (i.e., meso – diaminopimelic acid in the peptidoglycan together with arabinose and galactose in cell hydrolysates)distinguishing them from other groups of the same family15.Saccharomonospora azureais one of the 9 speciesof the genus Saccharomonospora14. Scientific reports on S.azureaare meagre with few reports on morphological characterization, physiology, genomics& secondary metabolite production and no significant studies on the nature of the pigment produced14,16–19.
This study aims to isolate and identify a pigment–producing actinomycete from rhizosphere soil followed by morphological and biochemical characterization of the isolate and its 16S rDNA molecular sequence & phylogeny analysis, so as to report a potential source for extraction of microbial pigment.
Materials and Methods
Sample Collection
Rhizosphere soil samples were collected from the gardens and pots at St. Joseph’s College (Autonomous), Bengaluru, Karnataka, India with a geographical location N:120 57’ 45.72”, E:770 35’ 49.56”. The samples were procured from a depth of 8 – 10cm using a sterile spatula and transferred to sterile glass bottles20.The samples were brought to the laboratory and processed further immediately.
Isolation and Screening of Actinomycetes
In order to isolate actinomycetes from different rhizosphere soil samples, 1g of the respective sample was suspended in 9ml of distilled water taken in a pre–autoclaved sterile test tube. The suspension was then serially diluted up to 10–5. The diluted suspension (100μl) of10–3,10–4, and 10–5was spread plated on Actinomycetes Isolation Agar (AIA) medium and incubated at 370 C for 3 ~ 7 days. AIA medium was prepared by dissolving2.17g AIA (Hi-Media) and 2g Agar–Agarin 100ml distilled water.100μl of Erythromycin and Fluconazole was added to the medium post autoclaving. Appropriate amount (30ml) of hot media was then poured into sterile petri plates and allowed to solidify at room temperature. Post incubation, all the plates were screened for actinomycetes colonies based on morphology and pigmentation. The actinomycete colony that showed significant coloration was sub–cultured onto fresh AIA medium by streaking until a pure culture was obtained.
Characterization of the Isolate
The isolate was characterized by studying its morphological (colony characters and Grams nature) and biochemical characteristics using standardized protocol. Colony characters such as form, margin, elevation, pigmentation, and texture were visually studied, and the observations were noted as described in Bergey’s Manual of Systemic Bacteriology20, 21. Grams nature of the isolate was determined using the standard protocol that uses Crystal Violet as the primary stain, an iodine solution as a mordant, 95 % ethanol as a decolourizer and Safranin as the counterstain22. Biochemical tests such as Carbohydrate fermentation, Simmons’ citrate (citrate utilization), Triple Sugar Iron (TSI) and Rapid Urease Test (RUT) were performed using standard protocols23-25.Glucose was used as the sole carbon source for the carbohydrate fermentation test. A lead acetate strip was used to detect H2S production for TSI test26.
Molecular Sequencing and Phylogeny analysis
The identity of the isolate was determined using 16S rDNA molecular sequencing analysis. The isolate was outsourced to Yaazh Xenomics, Coimbatore, Tamil Nadu, India for this purpose. The molecular identification technique was carried out as per the standard genome sequencing protocol. Genomic DNA was isolated using the EXpure Microbial DNA Isolation Kit developed by Bogar Bio Bee Stores Pvt. Ltd. The procedure for Genomic DNA isolation involved lysis, homogenization, centrifugation, binding (using binding buffer), washing and elution. The isolated Genomic DNA was amplified using PCR by following the regular thermal cycling conditions of denaturation, annealing and extension. Purification of the PCR products was done using the Montage PCR Clean Up Kit (Millipore) and PCR products were sequenced using primers with the help of the Terminator Cycling Sequencing Kit. Single pass sequencing was performed on each template using 16s rRNA universal primers. The fluorescent labelled fragments were purified using ethanol precipitation and the samples were resuspended in distilled water and subjected to electrophoresis in a sequencer27–29. The sequence obtained was subjected to BLAST analysis (https://blast.ncbi.nlm.nih.gov/) to identify its phylogenetic origin through homology. The generic and specific epithet of the strain was confirmed based on percentage similarity with the homologous sequences. The sequence data was submitted to the NCBI GenBank Database for public access. The most significant matches obtained from the NCBI GenBank Database (https://www.ncbi.nlm.nih.gov/genbank/) through BLASTn were utilized for constructing a phylogenetic tree under the Maximum Likelihood (ML) criterion using MEGA-X software.
Results
Isolation and Screening of Actinomycetes
Among all the rhizosphere soil samples collected, only one colony exhibited a powdery consistency with pigmentation. The suspected colony was obtained from a 10–3 serial dilution of a soil sample which was procured from the roots of weeds growing along with potted plants (Figure.1A).A pure culture was obtained by sub – culturing a loop full of the colony onto fresh AIA medium by streaking (Figure.1B).
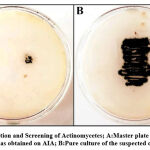 |
Figure 1: Isolation and Screening of Actinomycetes; A:Master plate with bacterial isolate as obtained on AIA; B:Pure culture of the suspected colony. |
Characterization of the Isolate
The morphological (colony characters and Grams nature) and biochemical characteristics of the isolate as observed have been summarized in Table.1. The colony had a filamentous margin and an elevation that appeared to be slightly raised. Prominent greenish–blue pigmentation was observed on the underside of the colony. The colony had a powdery consistency with a chalky azure aerial mass. On Gram staining the organism appeared to have gram–positive filamentous hyphae (Figure.2A). Biochemical analyses showed the organism to be positive for carbohydrate fermentation with no gas production (Figure.2B) and exhibited glucose fermentation only, with gas and H2S production for TSI test (Figure.2D). The organism tested negative for citrate utilization (Figure.2C) and RUT (Figure.2E).
Table 1: Morphological& biochemical properties of the pigment-producing actinomycetes train
| Sl. No. | Characteristics | Inference |
| 1 | Form | Filamentous |
| 2 | Margin | Filamentous |
| 3 | Elevation | Raised |
| 4 | Pigmentation | Greenish – blue pigmentation on the underside of the colony |
| 5 | Texture | Powdery with chalky azure aerial mass |
| 6 | Grams nature | Gram – positive |
| 7 | Carbohydrate fermentation | Glucose fermentation with no gas production |
| 8 | Citrate utilization | Negative |
| 9 | Triple sugar iron | Dextrose fermentation only, with gas (CO2 and H2) and H2S production |
| 10 | Urease | Negative |
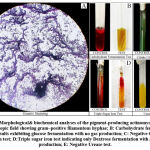 |
Figure 2: Morphological& biochemical analyses of the pigment-producing actinomycete strain. |
Molecular sequencing and Phylogeny analysis
The details of the16S rDNA molecular sequencing analysis have been summarized in Table.2. The length of the sequence was found to be 1307bp. The 1307bp sequence of the isolate was aligned using the BLAST n tool with pre–existing 16S rDNA gene sequences of microorganisms in the NCBIGen Bank database. The nucleotide alignment showed high similarity (100%) with S. azurea. The sequence data was submitted to the NCBI GenBank database under accession number MT509549 as ‘Saccharomonospora azurea strainSJCJABS01’. The phylogeny analysis of the sequence has been summarised in Figure.3. The phylogenetic tree constructed under the ML criterion using MEGA.X software highlights the position of S. azurea strain SJCJABS01 among its phylogenetic neighbours.
 |
Table 2: Sequencing analysis of 16S rDNA of isolated actinomycete |
 |
Figure 3: Phylogenetic treehighlighting the position of S. azurea SJCJABS01. |
Discussion
Biological pigments, especially from microbes are of great importance due to the some of the following reasons: microbes have rapid multiplication rates, they can be grown in low-cost media with ease, pigments can be processed easily with simple techniques when compared to plant pigments, etc. At present, there is high demand for microbial pigments in the global market because they are preferred over plant pigments and synthetic dyes due to their low production cost and eco – friendly nature respectively13. Due to the aforementioned reasons, the present study was aimed at isolating and identifying a pigment producing bacteria from rhizosphere soil. The main focus was to isolate a pigment–producing actinomycete because pigment production is more widely present in this group.
In attempts to isolate a pigment–producing actinomycete, a greenish–blue pigment–producing colony was isolated from one of soil samples by serial dilution method using AIA as the medium. The isolate had a filamentous margin, and an elevation that appeared to be slightly raised with prominent greenish–blue pigmentation on the underside of the colony. The colony exhibited powdery consistency with a chalky azure aerial mass. On Gram staining the organism appeared to have gram–positive filamentous hyphae. Biochemical analyses showed the organism to be positive for carbohydrate fermentation with no gas production and exhibited dextrose fermentation only, with gas and H2S production for TSI test. The identity of the isolate was confirmed using 16S rDNA and phylogeny analysis which revealed it to be Saccharomonospora azurea.
Conclusion
By definition, a pigment refers to any substance capable of absorbing light. Basically, it is the property of electromagnetic radiation with wavelengths ranging from 300–400nm and 700–800 nm30. During the 19th century, development of synthetic colors gained popularity due to their chemical stability, economical production cost and expansive spectrum of shades. However, subsequent discoveries enumerating the possible antagonistic effects of synthetic colorants like – allergenicity, carcinogenicity, hyperactivity, toxicity, etc., in humans and environmental pollution to name a few, demeaned their popularity leading to the increase in demand for natural alternatives. Microbial pigments, also referred to as biological pigments/ biochromes are simply pigments produced by microorganisms. A multitude of such pigments have been isolated and characterized from various microbes because of their wide–ranging bioactivity, for example: Phycoerythrin, Phycocyanin and Scytonemin from Cyanobacteria exhibit antioxidant, antitumor and immunoregulatory effects; Canthaxanthin from Haloferax alexandrines, Bradyrhizobium spp. and Lactobacillus pluvalis showcases anti–inflammatory, antioxidant, antitumor and photo–protectant activity; Anthraquinones from Penicillium oxalicumare antifungal and virucidal; while Melanin from Neoformans is an antibiofilm, antimicrobial and antioxidant. Even though many such microbial pigments may be rendered useless because of their instability against chemical and physical condition, various techniques have been developed to produce more stable pigments, for instance: microencapsulation, nano–emulsion formation and nano–formulations have been utilized to improve stability and deliverance of biological pigments to food matrices8. Moreover, pigments obtained from plants are limited and require complex production processes making them expensive in contrast to microbial pigments that are available in varying shades and can be easily produced in sufficient amounts with simpler processing techniques making them inexpensive2. Hence, due to these reasons pigment production in microorganisms is currently one of the most promising fields of research.
The present study was conducted so as to report a potential source for the extraction of microbial pigment. The main objective was to isolate and identify a pigment–producing actinomycete. This objective was driven by the existence of reports that are suggestive of pigment production occurring predominately in actinomycetes and the supporting evidence conclusive of their biological activities has proven these bacteria to be the quintessential targets for microbial pigments. A study conducted on Streptomycessp. showed that a melanin pigment produced through solid state fermentation is a potential source of antioxidants like tocopherol and troxol6. Another study on actinomycete pigment Prodigiosin from Streptomyces sp. revealed its antiproliferative property against Human Cervical Adenocarcinoma (HeLa) cell line, therefore, deducing actinomycetes to be extremely useful for pharmaceuticals3. An investigation undertaken by Parmar, R. S. & Singh, C., (2018) aimed at identification, characterization, and application of actinomycetes pigment showed that the isolate – actinomycete ARITM02, selected on the basis of its ability to produce a diffusible pigment was tested safe to be used as a natural food and pharmaceutical colorant. The conclusion of the study suggested that the novel versatile pigment was safe to be utilized in cosmetic, food, pharmaceutical and textile industries31. Miscellaneous research pertaining to aspects such as lip balm production and textile dyeing from actinomycete pigments exhibited prominent results for dye retention hence, making actinomycetes useful in cosmetic and textile industries2.
A powdery, greenish–blue colony with a chalky azure aerial mass was isolated on AIA from one of the many rhizosphere soil samples collected from our college campus. Upon preliminary investigation, viz. colony characterization and grams staining, the suspected colony was observed to have a filamentous margin with a slightly raised elevation and gram–positive filamentous hyphae. Biochemical analyses of the organism revealed positive results for carbohydrate fermentation and TSItest with no signs of gas production during the former but gas & H2S production during the latter. The isolate tested negative for citrate utilization and urease production. Identification of the isolate was achievedby 16S rDNA molecular sequence and phylogeny analysis. Results of the analysis strongly suggested that the pigment–producingsoil isolate was Saccharomonospora azurea. Correlations of morphology with previously reported S. azureas trains i.e., NA–128T14,16 and AP–11/1818 further substantiate our observations. Sparse research restricted tomorphology, physiology, genomics & secondary metabolite production with no reports on the physicochemical properties of the pigment produced by the identified organism clearly suggests that it is a relatively novel theme. Hence, further research pertaining to the nature of the pigment could be a benefactor to the scientific community.
Acknowledgement
The authors would like to thank St. Joseph’s College (Autonomous), Bengaluru, Karnataka, India for supporting and providing the facilities to carry out this study.
Conflict of Interest
No known conflict of interest for this work.
Funding Sources
No financial support was availed for this study.
References
- Amsaveni R, Sureshkumar M, Vivekanandhan G, Bhuvaneshwari V and Kalaiselvi M. Screening and isolation of pigment producing actinomycetes from soil samples. International Journal of Biosciences and Nanoscience 2015; 2: 24-28.
- TandaleA, KhandagaleM, PalaskarR and Kulkarni S. Lip balm production from pigment producing actinomycetes. IAETSD Journal for Advanced Research in Applied Sciences 2018; 5: 556-562.
- UdhayakumarK, RamalingamS, SaravananR and B D. Extraction of actinomycetes (streptomyces sp) pigment and evaluation of its anticancer property on HeLa cell line. Der Pharma Chemica 2017;9: 106–113.
- DastagerS, LiW.-J, Tian X.-P and Zhi X.-Y.Seperation, identification and analysis of pigment (melanin) production in Streptomyces Establishment of “National Culture Collection of Pakistan (NCCP)” View project Comparative study of Diversity, Molecular Phylogeny, and Bioactive Potential of Endophytes Associated with medicinal plants from Xinjiang and Uzbekistan View project 2014; 1131–1134.
- Sánchez-MarroquínA and Zapata M. Observations on the Pigment of Streptomyces coelicolor.Applied Microbiology1954; 2:102–107.https://doi.org/ 10.1128/aem.2.2.102-107.1954
CrossRef - DharmikP and GomasheA. Isolation, Identification and Antioxidant Activity of Melanin Pigment from Actinomycete (Streptomyces Species) Isolated from Garden Soil, Nagpur District, India. International Journal of Pure and Applied Sciences and Technology 2013; 18: 69–72.
- Greiner-maiE, Korn-WendischF and KutznerHJ. Taxonomic Revision of the Genus Saccharomonospora and Description of Saccharomonospora glauca sp. nov. International Journal of Systematic Bacteriology1988;38: 398–405. https://doi.org/10.1099/00207713-38-4-398
CrossRef - Sen T., Barrow C.J and Deshmukh S.K. Microbial Pigments in the Food Industry—Challenges and the Way Forward. Front. Nutr. 2019; 6:7. doi: 10.3389/fnut.2019.00007
CrossRef - SchuepW, BlountJ, WilliamsTandStempelA. Production of a novel red pigment, rubrolone, by Streptomyces echinoruber sp. NOV. II chemistry and structure elucidation. The journal of antibiotics 1978.
CrossRef - MuraseM, Hrkiji T, NittaK, TakeljchiYandUmezawaH. Peptimycin, a product of Streptomycesexhibiting apparent inhibition against ehrlich carcinoma. The journal of antibiotics, Ser. A 1960; 14: 113-118.
- BalagurunathanR, Selvameenal L and RadhakrishnanM. Antibiotic pigment from desert soil actinomycetes; biological activity, purification and chemical screening. Indian Journal of Pharmaceutical Sciences 2009; 71: 499. https://doi.org/10.4103/0250-474x.58174
CrossRef - R. P, Shobha R and OnkarappaR. Fascinating diversity and potent biological activities of actinomycete metabolites. Journal of Pharmacy Research 2010;3: 250–256.
- Lingappa K andNarasing RM. Biotechnological production of biocolor from microorganisms. Gulbarga university department of microbiology 2016; 1-110.https://doi.org/http://hdl.handle.net/10603/96411
- P, HeldB, LucasS, LapidusA, CopelandA, HammonN, Pitluck S, Goodwin L. A, HanC, Tapia R, Brambilla E.-M, PötterG, Land M, IvanovaN, RohdeM, GökerM, Detter J. C, KyrpidesN. C andWoykeT. Genome sequence of the soil bacterium Saccharomonospora azurea type strain (NA-128T). Standards in Genomic Sciences 2012; 6: 220–229. https://doi.org/10.4056/sigs.2635833
CrossRef - Syed D. G, Tang, S.-K, Cai M, Zhi X.-Y, AgasarD, LeeJ.-C, KimC.-J, Jiang C.-L, Xu L.-H and Li W.-J. Saccharomonosporasaliphila sp. nov., a halophilic actinomycete from an Indian soil. InternationalJournalOfsystemic and evolutionarymicrobiology 2008; 58: 570–573. https://doi.org/10.1099/ijs.0.65449-0
CrossRef - Runmao H. Saccharomonospora azurea nov., a New Species from Soil. International Journal of Systematic Bacteriology 1987;37: 60–61. https://doi.org/10.1099/00207713-37-1-60
CrossRef - Csepregi K, Valasek A, Pénzes A, Tóth Z, Írisz Kiss E, Kerepesi I, Horváth B, Nagy I and Feketea C. Draft Genome Sequence of an Efficient Antibiotic-Producing Industrial Strain of Saccharomonospora azurea, SZMC 14600. Journal of Bacteriology 2011; 1263.
CrossRef - Padhiar AR, Sharma MC and Modi HA. Isolation and preliminary optimization ofSaccharoromonospora azurea AP-11/18 for alkaline thermostable lipase production. Life sciences Leaflets 2012; 2: 94- 102.
- Kovács M, Seffer D, Pénzes-Hűvös Á, Juhász A, Kerepesi I, Csepregi K, KovácsValasek A and Fekete C. Structural and functional comparison of Saccharomonospora azurea strains in terms of primycin producing ability. World Journal of Microbioloy and Biotechnology 2020; 36, 160.
CrossRef - Nageswaran N, Ramteke P.W, Verma O.P and Pandey A. Antibiotic Susceptibility and Heavy Metal Tolerance Pattern of Serratia Marcescens Isolated From Soil and Water. Journal of Bioremediation and Biodegradation 2012;03. doi:10.4172/2155-6199.1000158
CrossRef - Bergey D.H, Staley J.T and Holt J.G. Bergey’s Manual of Systematic Bacteriology.Williams & Wilkins, 1989.
- Smith A.Cand Hussey M.A. Gram stain protocols. American Society for Microbiology 2005; 1:14.
- Atlas R.M. Handbook of Microbial Media for the Examination of Food.Crc;1995.
- Atlas R.M. Handbook of Media for Environmental Microbiology. Taylor & Francis, Second edition 2005.
CrossRef - Vasanthakumari R. Practical Microbiology. B.I. publications Pvt. Limited; 2009.
- Yi-Wei Tang, Stratton C.W. Advanced Techniques in DiagnosticMicrobiology. Springer,First edition 2006.
- Abirami M, and Kannabiran K. Antibiotic potency of extract from streptomyces isolated from terrestrial soil of Amirthi forest, India. Walailak Journal of Science and Technology 2017;14:711–721.
- Mani A, Ravi L and Krishnan K. Antibacterial and antifungal potential of marine streptomyces sp. Vitak1 derived novel compound pyrrolidinyl-hexadeca-heptaenone by in silico docking analysis. Research Journal of Pharmacy and Technology 2018;11:1901–1908.
CrossRef - Nabila M.I and Kannabiran K. Antagonistic activity of terrestrial streptomyces sp. Vitnk9 against gram negative bacterial pathogens affecting the fish and shellfish in aquaculture. Revista de Biologia Marina y Oceanografia 2018;53:171–183.
CrossRef - Herrera AS. The biological pigments in plant physiology. Agricultural sciences 2015; 6: 1262-1271.
CrossRef - Parmar RS and Singh C. A comprehensive study of eco-friendly natural pigment and its applications. Biochemistry and Biophysics reports 2018; 13: 22- 26.
CrossRef







