Rania Fawzy Mahmoud Abdelkawy1 , Shams Kholoussi1
, Shams Kholoussi1 , Eman Eissa1
, Eman Eissa1 , Khaled Hamed2
, Khaled Hamed2 , Haiam Abdel Raouf1
, Haiam Abdel Raouf1 * and Hala T. El-Bassyouni2
* and Hala T. El-Bassyouni2
1Immunogenetics Department, National Research Centre, Egypt
2Clinical Genetics Department, National Research Centre, Egypt
Corresponding Author E-mail: haiamabdelraouf@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/2236
Abstract
Background: Familial Mediterranean fever (FMF) is an auto inflammatory genetic disease resulted from the mutation of pyrin, which contributes to the formation of inflamma some complex. Therefore, activation of cytokines is one of the hallmarks of FMF pathogenesis. This study aimed to investigate the role of miRNAs as regulatory biomarkers for inflammation in patients with FMF. Methods: 50 FMF patients and 25 healthy subjects were included in this study. Q RT-PCR was used to determine plasma expressions of miR-181a and miR-125a, while IFN-γ and IL-17 were estimated using ELISA technique. Results: Our results indicated that, the expression of miR-181a was significantly decreased (p = 0.006) while miR-125a expression was insignificantly reduced (p = 0.101) also IL-17 levels were significantly higher(p = 0.003) and plasma IFN-γ levels were insignificantly increased (p = 0.322) in FMF patients than control group. Correlation analysis revealed a positive correlation between miR-181a expression and lymphocyte percentages (p = 0.048),while a significant negative association was observed between miR-125a and C-reactive protein (CRP) (p = 0.005) in FMF patients. However, there were no associations between miR-125a and miR-181a with IFN-γ and IL-17 in FMF patients. Conclusion: miR-181a and miR-125a could be used as regulatory biomarkers for inflammation in FMF patients.
Keywords
FMF; IFN-γ; IL-17; Inflammation; microRNAs
Download this article as:| Copy the following to cite this article: Abdelkawy R. F. M, Kholoussi S, Eissa E, Hamed K, Raouf H. A, El-Bassyouni H. T. Differential Expression of micro RNAs and their Association with the Inflammatory Markers in Familial Mediterranean Fever Patients. Biomed Pharmacol J 2021;14(3). |
| Copy the following to cite this URL: Abdelkawy R. F. M, Kholoussi S, Eissa E, Hamed K, Raouf H. A, El-Bassyouni H. T. Differential Expression of micro RNAs and their Association with the Inflammatory Markers in Familial Mediterranean Fever Patients. Biomed Pharmacol J 2021;14(3). Available from: https://bit.ly/3hUh2ON |
Introduction
Familial Mediterranean Fever (FMF) is an auto inflammatory disease commonly found among Eastern Mediterranean population. FMF occurred due to mutations in the MEFV (MEditerraneanFeVer) gene 1, 2. Incorrect coding resulted from MEFV mutations disturbs function of pyrin protein, and leads to uncontrolled inflammation. Several studies examining genotype-phenotype correlation in FMF patients with different clinical findings and therapeutic approaches showed that FMF is the paradigm of all the monogenic auto inflammatory disease. Pyrin is implicated in the formation of inflamma some complex. Pyrin impairment leads to auto inflammatory disease, resulting in aberrant production of interleukin (IL)-1β and IL-18. Consequently, cytokine activation is involved in the pathogenesis of FMF 3. Neutrophils play a major role in the inflammatory processes during the attacks of FMF. There are data showing persistent inflammation in attack-free MF patients as indicated by elevated levels of certain proinflammatory cytokines. 4-6. Interleukin (IL)-17 can modulate certain neutrophil functions by stimulatingtheir maturation andmigration. Elevated IL-17 leads to massive peripheral neutrophilia associated with increased levels of granulocyte colony stimulating factor (G-CSF) and enhanced granulopoiesis 7. It can recruit neutrophils into the peritoneal cavity by neutrophil-specific chemokines released from the peritoneal mesothelium 8. Amplification of persistent inflammatory responses may be the primary function of IL-17,as it can activatemany cell types as well as stimulate the secretion of several inflammatory cytokines including TNF-a, IL-6, IL-8, IFN-γ, and chemokines 9,10. Treatment with colchicine has been found to reduce these cytokines. Epigenetic pathways may be responsible for the variability of the clinical presentation of FMF such as miRNAs whichcould be a part of these pathways 11.
MicroRNAs (miRNAs) are small evolutionarily preserved non-coding RNA molecules (16-24 nucleotides) that disturb expression of their target mRNAs and have a role in biological processes as in cell growth, differentiation, and death. Diverse subsets of CD4+ T cell like Th1, Th2, Th17, and T regulatory cells, have several functions in immune activation and tolerance. Theyaredemonstrated to respond to dynamic micro-environmental indices and be involved inregulation of T cell development, survival and functions. Thus, miRNAsare implicated intheimmune physiological condition, on the one aspect, and havea role in controlling the immune tolerance, on the opposite. The cytokines are among the main proteins thatmiRNAstarget; these cytokinesserve as vital upstream signals and primary functional outputs 12.
It was suggested that, miRNAs can be used as a biomarker in various diseases. Plasma expression of miRNAs varies in many autoimmune and auto-inflammatory conditions. Consequently, miRNAs could have a regulatory function in the development and activation of inflammation and could be useful for diagnosing and monitoring the inflammation-related disorders 3.Furthermore, it has also been found that miRNAs are involved in the development of several immune cells, including development and proliferation of T and B lymphocytes, neutrophils, and regulate the release and activation of inflammatory mediators 11.
MiRNA-181 (miR-181) is preferentially expressed in many organs, especially in the bone marrow and spleen in significant levels. Due to its up-regulation in the spleen, miR-181 has been considered for its potential requirement and role in T-cell development and survival 13. Recent evidences have reported the implication of miR-181 members in the differentiation and functions of immune cells, such asdifferentiation and activation of B and T lymphocytes 14. Additionally, gene ontology investigation of predicted targets of miR-181a and -bhasdetected an over-representation of immune pathways involvingsignaling of T-cell receptor and transforming growth factor beta (TGF)-β 15.
MiRNA-125a (miR-125a) regulates expressions of several cytokinesimplicated in naive CD4 + T-cell differentiation in humans. The chronic expression of miR-125a decreased levels of cytokines likeIL-10 receptor α, IL-2 receptor β, andIFN-γ. Introducing miR-125 into naive CD4+ T-cells resulted inreducingexpressions of molecules presented on the surface of Th1 and Th2 memory cells and led toelevation of naive CD45RA+, CD45RO-, elucidating miR-125 function in the preservation of tolerance and keeping naive T-cell status. In addition, miR-125 may reduce the effector function and activationof T-cells, which is shown by decreased levels of intracellular IFN-γ and IL-13. MiR-125 down-regulation is accompaniedby effector memory CD4+ T-cell phenotype 14.
In the current study, we object to investigate the potential involvement of microRNAs in the regulation of inflammation in FMF patients. Also, to examine the expression patterns of miR-125a and miR-181a and plasma levels of the inflammatory cytokines (IFN-γ and IL-17) in FMF patients compared to healthy controls. In addition, we evaluate the correlation between these miRNAs and the clinical and laboratory manifestations of FMF patients,and then estimate their association with IFN-γ and IL-17 expression in FMF patients.
Patients and Methods
Ethics
Ethics Committee of the National Research Center (NRC), Giza, Egypt, approved this study and written informed consents were obtained from the parent/guardian of all children at enrolment and before any study procedure.
Study Subjects
In this case–control study, 75 subjects were included, with age ranging from 3 to 16 years. There were 50 patients with FMF and 25 apparently normal controls.
Patients were recruited from the Clinical Genetics Department, Medical Research Center of Excellence, National Research Centre, Giza, Egypt.Patients with FMF were diagnosed according to the Tel Hashomer Diagnostic Criteria [16], and were on colchicine treatment at the time of the study.Clinical Characteristics and treatments of FMF patients are shown in table 1.
RNA extraction and Reverse Transcription
MiRNAs were isolated and extracted from plasma of all subjects of the study groups using miRNeasy Mini kit (Qiagen, Germany) and by following the manufacturer’s instructions.cDNA was synthesized usingTaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems) and using specific primers by following the manufacturer’s instructions. Reverse transcription was performed under the following thermal conditions: starting at 16 °C for 30 min followed by 42 °C for 30 min and finally at 85 °C for 5 min and the resulting cDNA was kept at −80 °C until use.
Real time PCR quantification
A real-time quantitative PCR (qRT-PCR) was doneusing TaqMan® MicroRNA Assay kit and TaqMan® Universal Master Mix (Applied Biosystems) to quantify the expression levels in triplicate of mature miR-181a and miR-125a using 7500 fast real-time PCR system by following the manufacturer’s instructions. RNU48 was used as a reference gene (housekeeping gene). A single plex reaction was used in this study. The expression levels of target miRswere normalized toRNU48 and relative quantification (Rq) of miRNA expression was calculated using RQ formula(2−ΔΔCT). ΔCt was determined by subtracting the Ct values for RUN48 from the Ct values for the target miR. Q RT-PCR was carried out with cycling conditions of: 50°C for 2 min, 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and1 min at 60°C 17.
Determination of IFN-γ andInterleukin 17
Plasma IFN-γ and IL-17 levels of study patients and healthy subjects were assessed in duplicate by the “commercially-available solid-phase sandwich ELISA kit (ELISA)”(Elabscience, Elabscience Biotechnology Co., Ltd) following the protocol provided by the manufacturer.
Data analysis and statistics
Data were collected, revised, verified then analyzed using SPSS version 19.0 software (SPSS Inc., Chicago, Illinois, USA). Comparison between expression levels of miRs and cytokines was performed usingnon-parametric Mann-Whitney U test.Spearman¢s rank correlation to test the association of miR expression levels with laboratory data and inflammatory cytokines of patients. P value of <0.05 was reflected in statistical significance.Quantitative data were described using mean and SD or median and range.
Results
Demographic and clinical data
Demographic and clinical characteristics as well as laboratory findings of 50 patients with FMF are presented in Table (1).
Table 1: Clinical and laboratory findings among FMF group
| Characteristic | FMF patients(n=50) | |
| Gender, male/female % | 52/48% | |
| Age, median (years) (Range) | 9.5 (3-16) | |
| Consanguinity % | 42% | |
| Family history % | 66% | |
| Inflammatory attack, positive/negative % | 58/42% | |
| Colchicine Responders/ Non-Responders % | 78/22% | |
| Medications (colchicine*) % | 100% | |
|
MEFV genotype, % of patients |
||
| Homozygous | M680I | 14% |
| M694I | 8% | |
|
Heterozygous |
M694I | 46% |
| V726A | 10% | |
| E148Q | 18% | |
| R761H | 4% | |
| Laboratory findings | ||
| Hemoglobin(g/dl), Mean ± SD (Range) | 13 ± 1(11.3-15.7) | |
| Platelets (× 103/mm3), Mean ± SD (Range) | 280±97(50-450) | |
| WBCs (× 103/mm3), Median (Range) | 6.8(4.7-22.9) | |
| Lymphocytes%, Mean ± SD (Range) | 47.5±10.3(29-64) | |
| Neutrophils%, Mean ± SD (Range) | 43±10(28-63) | |
| CRP (mg/L), Median (Range), (normal ≤ 5) | 16 (1.4-45) | |
| ESR (mm/hr), Median (Range) | 24 (5-56) | |
* The dose of colchicine ranged from 0.5-1.5 mg/day
Evaluation of plasma miR-181a and miR-125a expression
Our findings delineated that,miR-181a expression was significantly down-regulated in FMF patients in comparison with healthy controls. In FMF patients, miR-181a expression level was 5.85-fold lower than healthy controls (Table 2).
In addition, our results demonstrated the under-expression of miR-125a in patients with FMF in comparison the control group. 2.27-fold down-regulation of miR-125a expression was detected in FMF patients in comparison with control group (Table 2).
Table 2: Expression of miR-181a and miR-125a in FMF patients
| N | Rq Median (Min-Max) | P value | |
| miR-181a | 50 | 0.171 (0.01-1.2) | 0.006* |
| miR-125a | 50 | 0.44 (0.04-2.08) | 0.101 |
Rq: The Relative Quantification
* Significant atP< 0.01 compared with controls using Mann-Whitney test.
Plasma levels of inflammatory cytokines
Our results showed that, the levels of IFN-γ were higher in FMFpatients (Median, Range: 108, 33-263) than that of the control group (Median, Range: 81, 35-191) (P=0.322). Moreover, IL-17 expression was significantly elevated in FMF patients (Median, Range: 11, 3.77-92.74) compared with healthy controls(Median, Range: 2.9, 0.34-8.41)(P=0.003) (Figure1).
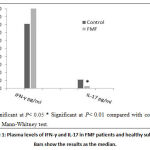 |
Figure 1: Plasma levels of IFN-γ and IL-17 in FMF patients and healthy subjects. Bars show the results as the median. |
The correlation of the plasma miR-181a and miR-125a expression with the laboratory parameters and cytokineslevels in patients group
Correlation analysis showed that, miR-181a expression have a significant positive correlation with lymphocyte percentages in FMF patients while no correlation was detected with the other laboratory data (white blood cells, neutrophils, CRP and ESR) of FMF patients. Furthermore,our data indicated thatmiR-125a expression have a significant negative correlation with CRP while no correlations were found between miR-125a and the other laboratory data of FMF patients (Table 3).
In addition, our data indicated that there are no associations between miR-181a and miR-125awith the inflammatory cytokines (IFN-γ and IL-17) in FMF patients (Table 3).
Table 3. Correlation of miR-181a and miR-125a expression levels with the laboratory parametersand cytokines in FMF group
| miR-181a expression | miR-125a expression | |||
| R | P value | R | P value | |
| White blood cells (WBCs) | 0.486 | 0.154 | -0.394 | 0.260 |
| Lymphocytes | 0.636 | 0.048* | 0.419 | 0.228 |
| Neutrophils | -0.433 | 0.211 | -0.262 | 0.464 |
| CRP | -0.872 | 0.054 | -0.975 | 0.005* |
| ESR | -0.564 | 0.322 | -0.667 | 0.219 |
| IFN-γ | 0.261 | 0.467 | 0.212 | 0.556 |
| IL-17 | 0.552 | 0.098 | 0.382 | 0.276 |
R: Spearman’s correlation coefficien
*: significant correlation at the 0.05 level.
Discussion
FMF is an inherited autoinflammatory disease caused bythe pyrin mutation; this mutationparticipates in inflammasomecomplex formation. Consequently, activated cytokinescontribute to the pathogenesis and activation of FMF 18.
MiRNAs are small endogenous RNAs whichregulate gene expression post-transcriptionally by binding totheir targets. Recent studies have shown aberrant miRNA expression in various diseases such as the auto inflammatory diseases which indicated their potential effects in the pathogenesis of these diseases 3, 19. In current study, we aimed to assess the plasma expression of some candidate miRNAs associated with autoimmune pathogenesis and inflammation.
Our findings showed a significant down-regulationin the expression of miR-181a (-5.85 folds)of FMF patients compared with healthy subjects. Similar findings have been reported bya study of Hortuet al.(2019),in which there is a reduction in expression ofmiR-181a in FMF casesrelative to healthy controlswhile it was observed to have elevated when compared with patients not receiving colchicine therapy 11.
On the other hand, Karpuzogluet al. (2020) revealed that miRNAs expression levels were altered in the serum of patients with FMF when compared withthe control group. In detail, miR-181a, miR-181b, miR-181c, and miR-365a were deregulated. These miRNAs were suggested to target different genes, and upregulation of these non-coding miRNAs wasassociatedwith oncogenesis and autoinflammatory diseases, including FMF [20].Also, Hortuet al. (2019)worked on miRNAs more comprehensively in 51 pediatric FMF patients.They demonstrated that only 15 miRNAs including miR-181a and miR-125a had aberrant expressions 11.
Moreover, a study by Lashineet al. (2011) showed miR-181a under-expression in juvenile SLE patients. However, they described no difference between the healthy controls and FMF patients in that study 21.
In addition, our results demonstrated the under-expression of miR-125a (-2.27-fold) in FMF patients compared to the control group. This finding is consistent with the demonstration ofHortuet al. (2019) that the expression pattern of 11 miRNAs,including miR-125a and miR-181a, in the pediatricFMF patientswere markedly lowerthan those of the healthy control 11. Others demonstrated that miR-125a expression down-regulated in FMFpatients relative to the healthy control group 22.
It has been found that both miR-181a and miR-181c playimportant roles in premature stages of T cell development. miR-181a, which has a function in differentiation and activation of T-cells, is present in enough amounts during maturation of T-cells before CD4+ and CD8+ stages and is diminished in the later stages. TCR signaling after transcription is regulated by miR-181a, in which processup-regulation of miR-181a leads toelevation of TCR signaling in T-cells and vice versa[14]. That matches our study in whicha significant positive association between miR-181a expression level and lymphocyte percentages in FMF group.Similar results have been reported by Li et al. (2007) and Schaffertet al. (2015) who have shown that high levels of miR-181a and -b in various stages of T cell development causes induction of positive and negative choice via promoting TCR sensitivity and signaling strength in human and mice, respectively 23, 24.
Several studies [25,26]demonstrated that miR-125a has a marked role as an anti-inflammatory agent in controllingthe autoimmune diseases;this is in agreement with our results which indicated a significant negative association between miR-125a expression level and CRP of FMF group. This is in accordance with Murata et al [25] who found that miR-125a was negatively correlated with some indices of disease activity including CRP in Rheumatoid arthritis. Also, Sun et al [26]reported that miR-125a was negatively associated with CRP, ESR, IL-17, and TNF-α in Crohn’s disease patients.
In this study, we found that IL-17 expression was significantly up-regulated in FMF patients in comparison with healthy controls. In an earlier study performed by Koga et al. (2016), IL-17 and IL-18 levels inthe serum of FMF patients were markedlyincreases in comparison with those of healthy controlswhile they were comparable in FMF patients in attack and remission [18].Moreover, FMF patients have been reported by Kogaet al. (2018) to show an elevated level of serum inflammatory cytokines like IL-1β, IL-6, IL-17, and IL-18. They recently revealed the specific cytokine network amongst FMF patients through the use of a multi-suspension cytokine array 19. A previous study reported that the serum concentration of IL-17 is significantly increased during FMF attacks 27.
In this study, we found that the IFN-γ serum levels were higher in FMF patients than that of the control group. That is in agreement with Köklüet al.(2005) who showed that median IFN-γ plasma levels in FMF patients both with and without attack were significantly higher than the healthy controls(P < 0.05). In addition, higher IFN-γ plasma levels were observed in patients with acute FMF attacks in comparison with patients in attack-free periods (P < 0.05). IFN-γ plasma levels were comparable in colchicine treated and untreated patients 28.
Furthermore, our data indicated no associations between miR-125a or miR-181a with the inflammatory cytokines (IFN-γ and IL-17) in FMF patients. But it was found several other miRNAs that could be involved in the regulation of IL-23/IL-17 axis by indirect mechanisms in autoimmune diseases [29]. Other studies demonstrated that miR-125a and miR-181a were negatively correlated with IL-17 levels in patients with the active inflammatory disease [26, 30, 31].While it has been found that miR-181a expression is inverselyassociated with the severity of inflammation, it is still unclear which pathway this expression gets along 11.
In conclusion, miR-181a and miR-125a could be used as regulatory biomarkersfor inflammation in FMF patients. Further functional researches may be helpful to elucidate and clarify the role of miRNAs, especially in the regulation of inflammation in FMF. MiRNAsmight havea promising therapeutic roleinauto-inflammatory diseases as FMF.
Acknowledgement
We thank the National Research Centre (in-house office for research projects) for the research grants supported this work
Conflict of interest
The authors declare that they have no conflict of interest.
Funding Sources
This study was supported by National Research Centre, Giza, Egypt.
References
- Kishida D, Yazaki M, Nakamura A, Nomura F, Kondo T, Uehara T, Ikusaka M, Ohya A, Watanabe N, Endo R, Kawaai S. One novel and two uncommon MEFV mutations in Japanese patients with familial Mediterranean fever: a clinicogenetic study. Rheumatology international. 2018;38:105-10.
CrossRef - Sarrauste de Menthière C, Terrière S, Pugnère D, Ruiz M, Demaille J, Touitou I. INFEVERS: the Registry for FMF and hereditary inflammatory disorders mutations. Nucleic Acids Res. 2003; 31:282-285.
CrossRef - Demir F, Cebi AH, Kalyoncu M. Assessment of Circulating Microribonucleic Acids in Patients with Familial Mediterranean Fever. Archives of Rheumatology. 2020;35:52-59
CrossRef - Skendros P, Papagoras C, Mitroulis I, Ritis K. Autoinflammation: Lessons from the study of familial Mediterranean fever. J Autoimmun. 2019;104:102305.
CrossRef - Direskeneli H, Ozdogan H, Korkmaz C, Akoglu T, Yazici H. Serum soluble intercellular adhesion molecule 1 and interleukin 8 levels in familial Mediterranean fever. J Rheumatol. 1999;26:1983-1986.
- Kiraz S, Ertenli I, Arici M, et al. Effects of colchicine on inflammatory cytokines and selectins in familial Mediterranean fever. ClinExpRheumatol. 1998;16:721-724.
- Schwarzenberger P, La Russa V, Miller A, et al. IL-17 stimulates granulopoiesis in mice: use of an alternate, novel gene therapy-derived method for in vivo evaluation of cytokines. J Immunol. 1998;161:6383-6389.
- Witowski J, Pawlaczyk K, Breborowicz A, et al. IL-17 stimulates intraperitoneal neutrophil infiltration through the release of GRO alpha chemokine from mesothelial cells. J Immunol. 2000;165:5814-5821.
CrossRef - Gaffen SL. Biology of recently discovered cytokines: interleukin-17–a unique inflammatory cytokine with roles in bone biology and arthritis. Arthritis Res Ther. 2004;6:240-247.
CrossRef - Aggarwal S, Gurney AL. IL-17: prototype member of an emerging cytokine family. J Leukoc Biol. 2002;71:1-8.
- Hortu HO, Karaca E, Sozeri B, Gulez N, Makay B, Gunduz C, Atik T, Tekin IM, Unsal SE, Cogulu O. Evaluation of the effects of miRNAs in familial Mediterranean fever. Clinical rheumatology. 2019;38:635-43.
CrossRef - Garavelli S, De Rosa V, de Candia P. The multifaceted interface between cytokines and microRNAs: An ancient mechanism to regulate the good and the bad of inflammation. Frontiers in immunology. 2018;9:3012.
CrossRef - Feng MJ, Shi F, Qiu C, Peng WK. MicroRNA-181a, -146a and -146b in spleen CD4+ T lymphocytes play proinflammatory roles in a murine model of asthma. 2012; 13:347-53.
CrossRef - Saki N, Abroun S, Soleimani M, Hajizamani S, Shahjahani M, Kast RE, Mortazavi Y. Involvement of microRNA in T-cell differentiation and malignancy. International journal of hematology-oncology and stem cell research. 2015;9:33-49.
- Ghorbani S, Talebi F, Chan WF, Masoumi F, Vojgani M, Power C, Noorbakhsh F. MicroRNA-181 variants regulate T cell phenotype in the context of autoimmune neuroinflammation. Frontiers in immunology. 2017;8:758:1-14.
CrossRef - Livneh A, Langevitz P, Zemer D, et al. Criteria for the diagnosis of familial Mediterranean fever. Arthritis Rheum. 1997;40:1879-1885.
CrossRef - Elnady HG, Abdelmoneam N, Eissa E, Hamid ERA, Zeid DA, Abo-Shanab AM, Atta H, Kholoussi NM. MicroRNAs as Potential Biomarkers for Childhood Epilepsy. Open Access Maced J Med Sci.2019;7:3965-3969.
CrossRef - Koga T, Migita K, Sato S, Umeda M, Nonaka F, Kawashiri SY, Iwamoto N, Ichinose K, Tamai M, Nakamura H, Origuchi T. Multiple serum cytokine profiling to identify combinational diagnostic biomarkers in attacks of familial Mediterranean fever. Medicine. 2016;95:1-8.
CrossRef - Koga T, Migita K, Sato T, Sato S, Umeda M, Nonaka F, Fukui S, Kawashiri SY, Iwamoto N, Ichinose K, Tamai M. MicroRNA-204-3p inhibits lipopolysaccharide-induced cytokines in familial Mediterranean fever via the phosphoinositide 3-kinase γ pathway. Rheumatology. 2018;57:718-26.
CrossRef - Karpuzoglu EM, Ekinci RM, Balci S, Bisgin A, Yilmaz M. Altered expression of apoptosis-related, circulating cell-free miRNAs in children with familial Mediterranean fever: a cross-sectional study. Rheumatology International. 2021;41:103-111.
CrossRef - Lashine YA, Seoudi AM, Salah S, Abdelaziz AI. Expression signature of miRNA 181-a reveals its crucial role in the pathogenesis of paediatric systemic lupus erythematosus. ClinExpRheumatol 2011;29:351–357.
- Balci-Peynircioglu B, Akkaya-Ulum YZ, Akbaba TH, Tavukcuoglu Z. Potential of miRNAs to predict and treat inflammation from the perspective of Familial Mediterranean Fever. Inflammation Research. 2019;1:1-9.
CrossRef - Schaffert SA, Loh C, Wang S, Arnold CP, Axtell RC, Newell EW, et al. miR-181a-1/b-1 modulates tolerance through opposing activities in selection and peripheral T cell function. J Immunol. 2015;195:1470–9.
CrossRef - Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007;129:147–61.
CrossRef - Murata K, Furu M, Yoshitomi H, et al. Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS One. 2013;8:e69118:1-10.
CrossRef - Sun CM, Wu J, Zhang H, Shi G, Chen ZT. Circulating miR-125a but not miR-125b is decreased in active disease status and negatively correlates with disease severity as well as inflammatory cytokines in patients with Crohn’s disease. World J Gastroenterol.2017;23:7888-7898.
CrossRef - Ashida M, Koike Y, Kuwatsuka S, Ichinose K, Migita K, Sano S, Utani A. Psoriasis‐like lesions in a patient with familial Mediterranean fever. The Journal of dermatology. 2016;43:314-7.
CrossRef - Köklü S, Öztürk MA, Balcı M, Yüksel O, Ertenli I, Kiraz S. Interferon-gamma levels in familial Mediterranean fever. Joint Bone Spine. 2005;72:38-40.
CrossRef - Salvi V, Gianello V, Tiberio L, Sozzani S, Bosisio D. Cytokine targeting by miRNAs in autoimmune diseases. Frontiers in immunology. 2019;10:15:1-10.
CrossRef - Ye S, Zhu S, Feng L. LncRNA ANRIL/miR‐125a axis exhibits potential as a biomarker for disease exacerbation, severity, and inflammation in bronchial asthma. Journal of Clinical Laboratory Analysis. 2019;e23092:1-8.
CrossRef - Cao Y, Zhao D, Li P, Wang L, Qiao B, Qin X, Li L, Wang Y. MicroRNA-181a-5p impedes IL-17-induced nonsmall cell lung cancer proliferation and migration through targeting VCAM-1. Cellular Physiology and Biochemistry. 2017;42:346-56.
CrossRef








