Lokesh Singh , Rekh Ram Janghel
, Rekh Ram Janghel and Satya Prakash Sahu
and Satya Prakash Sahu
National Institute of Technology, G.E Road, Raipur, India.
Corresponding Author E-mail: lsingh.phd2017.it@nitrr.ac.in
DOI : https://dx.doi.org/10.13005/bpj/2225
Abstract
Purpose:Less contrast between lesions and skin, blurriness, darkened lesion images, presence of bubbles, hairs are the artifactsmakes the issue challenging in timely and accurate diagnosis of melanoma. In addition, huge similarity amid nevus lesions and melanoma pose complexity in investigating the melanoma even for the expert dermatologists. Method: In this work, a computer-aided diagnosis for melanoma detection (CAD-MD) system is designed and evaluated for the early and accurate detection of melanoma using thepotentials of machine, and deep learning-based transfer learning for the classification of pigmented skin lesions. The designed CAD-MD comprises of preprocessing, segmentation, feature extraction and classification. Experiments are conducted on dermoscopic images of PH2 and ISIC 2016 publicly available datasets using machine learning and deep learning-based transfer leaning models in twofold: first, with actual images, second, with augmented images. Results:Optimal results are obtained on augmented lesion images using machine learning and deep learning models on PH2 and ISIC-16 dataset. The performance of the CAD-MD system is evaluated using accuracy, sensitivity, specificity, dice coefficient, and jacquard Index. Conclusion:Empirical results show that using the potentials of deep learning-based transfer learning model VGG-16 has significantly outperformed all employed models with an accuracy of 99.1% on the PH2 dataset.
Keywords
CAD; Deep Learning; Inception; Melanoma; Skin Lesion; Transfer Learning; Xception
Download this article as:| Copy the following to cite this article: Singh L, Janghel R. R, Sahu S. P. A Deep Learning-Based Transfer Learning Framework for the Early Detection and Classification of Dermoscopic Images of Melanoma. Biomed Pharmacol J 2021;14(3) |
| Copy the following to cite this URL: Singh L, Janghel R. R, Sahu S. P. A Deep Learning-Based Transfer Learning Framework for the Early Detection and Classification of DermoscopicImages of Melanoma. Biomed Pharmacol J 2021;14(3). Available from: https://bit.ly/3jPFwJc |
Introduction
Early detection is vital which can improve the prognosis of individuals to be suffered from malignant melanoma1. Prognosis starts at first step with visual screening, at the second step, the dermoscopic investigation is performed, and at the third step, a biopsy is conducted which is proceeded by histopathological analysis2. Since,the melanoma lesion differs in appearance, shade, and areas for various types of skin3, investigation of dermoscopic images is vital in the prognosis of melanoma at a very early stage45. Several studies are in progress towards the classification of pigmented skin lesion images using various learning paradigms. The use of machine learning methods to classifylesion images requires prior knowledge and solely based on hand-crafted features that pose complexity. In a computer vision competition namely Image Net Large Scale Visual Recognition Challenge (ILSVR), an error rate of 30% has been observed using machine learning classifiers for the classification of around two million images into thousand categories 6. While the same competition was organized in 2012, an error rate of 26–30% is obtained using conventional machine learning methods which is then reduced to 16.4% when image classification is performed using convolutional neural network (CNN), a deep learning classifier7. The error rate is reduced drastically to below 5% using the deep learning architectures in the ILSVR competition organized in 20178. Numerous CNN pre-trained architectures are employed for image classification namely, LeNet9, AlexNet10, ZFNet11, VGGNet12, GoogLeNet13, ResNet14 etc. Pre-trained models have a higher capability of extracting features from the image using transfer learning. Despite many images using augmentation, thedataset’s sizeis sometimes not sufficient for training a model since scratch. The issue overcomes using transfer learning with the help of the pre-trained CNN architectures that canenhance the classification accuracy of pigmented skin lesion images by skilled dermatologists.However, the analysis of melanoma by experienced dermatologists often fail due to some of the difficultiessuch as the color of hairs, different areas, shade and appearance of a mole, the variance between the actual normal skin and lesion. Theserequirea computer aided diagnostic (CAD) system to help experts in classifyingthe lesion as benign or malignant.
In this work anautomated computer-aided diagnosis system for melanoma detection (CAD-MD) is designed for the effective and accurate classification of pigmented skin lesions as benign or malignant.CAD-MD is designed to assists dermatologists or medical experts in identifying the lesions The potentials of proposed system as follows:
An automated CAD-MD system is designed for the early and accurate detection and classification of melanoma as benign or malignant.
Image-downsizing is performed for reducing complexity that occurs during training and testing of the images.
Hair-noise removal is conducted using the digital hair removal (DHR) method.
For the easier analysis of image, significant region of interest (SROI) are extracted from the pigmented lesion images by performing segmentation using the Watershed algorithm.
Gray-level co-occurrence matrix (GLCM) is used for the better extraction of features from the segmented pigmented skin lesion image.
Two set of experimentations are carried with actual images and augmented images on dermoscopic PH2 and ISIC-2016 datasets.
The classification of pigmented lesions is carried out utilizing the machine learning, and deep learning-based transfer learning models with tuned parameters on actual and augmented images.
The performance of CAD-MD is evaluated on PH2 and ISIC-2016 dermoscopic datasets based on accuracy, precision, recall, f1-score and error-rate.
The organization of the paper is as follows- previous literature regarding the classification of dermoscopic skin lesion images is discussed in section-2 in brief. Section-3 elaboratesthe proposed CAD-MD system comprising the preprocessing, segmentation, feature extraction and classification methods. Performance measures are demonstrated in section-4. Experimental results are reported in section-5, while the summarized discussion and theconclusion of the work is demonstrated in section-6 and section-7.
Related Work
Deep CNN architecture is used 3 for extracting the skin lesion from the images. Overall, the proposed methodology gives a promising accuracy of 92.8%. However, combining the features obtained from several pretrained CNN networks may yield better-classified results. A methodology is proposed in15that performs a comparison between 1- level classier and 2-level classifier. The best performance results are achieved from a two-level classifier with promising accuracy of 90.6%. A novice classification system16is designed using the KNN technique for classifying the skin lesion. The designed system has achieved an accuracy of 93% during the experimentation. However, the system lacks significant improvement due to smaller size of the training set. An automated system17 isdesigned using deep learning methodology to detect melanoma and classified the dermoscopic image as benign or malignant. They proposed a Synergic Deep Learning (SDL) model using the Deep Convolutional Neural Network (DCNN) for addressing the challenging issues caused by the intra-class-variation and inter-class-similarity to classify skin lesions. They achieved an accuracy of 85.8% which is not quiet promising compared to the state-of-art-methods discussed in the work.A new approach for segmentation and classification18 isdesigned of the skin lesions as well as a Region growing technique forperforming the segmentation of extracting lesion areas. Extracted features are then classified by employing SVM and KNN classifiers and performance is measured using F-measure with 46.71%. A number of ongoing work towards designing an automated pigmented skin lesion classification system utilizing deep and transfer learning might save the medical expert’s time and effort.One such CAD is designed by 19,usinghand-crafted features based on color, shape and texture to combine those features with deep learning features towards detecting melanoma.Similar work had also been reported by20utilizing transfer learning. Another lesion classification system is designed by 21 usingthe pre-trained model’s deep convolutional network. Other frameworks have also been proposed for the automated classification of skin lesion images utilizing convolutional neural networks by 22, 23, 24. Several studies towards melanoma detection and classification at an early stage have also been reported in 25–29.Worldwide several researchers are working towards solving an un-balanced dataset for the classification of melanoma as benign or malignant. One such comprehensive work regarding solving the issues of skewed distribution has been conducted by30by employing several oversampling methods including ADASYN, ADOMS, AHC, Borderline-SMOTE, ROS, Safe-Level-SMOTE, SMOTE, SMOTE-ENN, SMOTE-TL, SPIDER and SPIDER2, and under-sampling methods including CNN, CNNTL, NCL, OSS, RUS, SBC, and TL. They investigated the effect of skewed class distribution over the learners employed and identified the best rebalancing method for several used cancer datasets. Instead of using conventional data-level balancing approaches, 22 have rebalanced the dataset by creating synthetic dermatoscopic lesion images from the under sampled class employing un-paired image to image translation. 31 has investigated the impact of an imbalanced dataset towards classifying the melanoma as benign or malignant by employing oversampling strategies including random oversampling (ROS), and SMOTE while they used Tomeklink(TL), random undersampling (RUS), neighborhood cleaning rule (NCR) and NearMiss undersampling methods for balancing the dataset. They also used hybrid sampling methods for dataset rebalancing, including SMOTE+TL and SMOTE+ENN.
Limitation of the related work
The functionalities of the conventional CAD system discussed in the literature, includes pre-processing, data augmentation, segmentation, feature extraction, and classification which assist the medical experts in detecting and interpreting the disease effectively. The majority of the literature lacks in designing such an effective system towards a cost-effective diagnosis of the disease where medical experts are not available. The problem of data limitation and skewed class distribution in dermoscopic datasets aggravates the issue in melanoma detection. The majority of the literature has focused more on classification instead of performing classification by solving the imbalance issues. Few studies reported in the literature have worked towards dataset rebalancing. Among several approaches towards balancing the uneven class distribution to skewed datasetsdata-level approaches are frequently used.It can be observed from the literature that the reported work rebalances the class distribution of the image dataset using oversampling and undersampling methods. However, oversampling methods generateduplicate samples from the minority class, which results inoverfitting and leads to other issues like class-distribution shift during several iterations. Another approach that has been frequently used in the literature for balancing the dataset is the use of under sampling methods. Though the under sampling methods balance the class distribution they results in information loss as they eliminate the samples from the majority class until the dataset gets balanced.
Our proposed CAD-MD system overcome the issue of data limitation and class skewness by utilizing image augmentaion techniques which artificially enhances the size of dataset without changing the semantic meaning of actual images while overcoming the class skewness. Using the data augmentation operations like rotation, vertical- horizontal flips, horizontal-vertical shear we have generated a new training dataset by transforming the images from the current lesion images belonging to the same class as the actual lesion images. The method minimizes the chance of overfitting and enhances the size and quality of lesion images.
Proposed Cad-Md System
This section discusses the proposed CAD-MD system designed for the effective and accurate detection and classification of pigmented skin lesion as benign or malignant. The framework of designed CAD-MD system is shown in Figure 1 as follows:
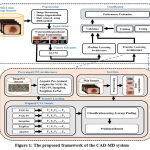 |
Figure 1: The proposed framework of the CAD-MD system. |
Dataset Acquisition
PH2 dataset 1 comprises Dermoscopy images which were obtained from the Dermatology Service of Hospital Pedro Hispano, Matosinhos, Portugal. The PH2 database comprises several criteria: colors, pigment network, dots/globules, streaks, regression areas, and blue-whitish veil assessed by the expert dermatologist doctors. Another dataset, ISIC-16 232, is a collection of some publicly available high-quality dermoscopic images of skin lesions. Figure 2 presents the samples of pigmented lesions of PH2 and ISIC datasets.
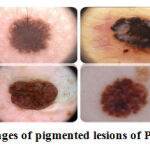 |
Figure 2: Sample images of pigmented lesions of PH2 and ISIC dataset. |
Preprocessing
Preprocessing of images is two fold
Image-Downsizing
At first, the size of the images is transformed. The original size of PH2 and ISIC-16 images dataset is 765 × 572 and 1022 × 767 respectively. These images were downsized to a size of 120 × 160 to reduce the complexity and computational time, consumed during training and testing.
Hair Removal
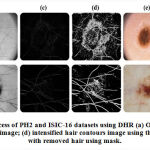
The images comprising hair-noise in PH2 and ISIC-16 datasets might affect the accuracy, which is being removed by employing DHR (Digital Hair Removal) algorithm. At first, the original-colored image is converted into a grayscale image. Then to find the hair contours, morphological blackhat filtering is applied on the gray scale image, where hair contours are then intensified towards preparing for inpainting. Finally, a hair-free image is obtained after applying inpainting on the original image as per the mask. The algorithm then starts from the region’s boundary and inpaints the region of an image by chosing a pixel near to the region’s pixel to be inpainted. It replaces the pixel to be inpainted by normalized weighted sum of neighboring pixels. The pixels lying closer to the region are provided higher weights. Once the pixel is inpainted,the algorithm moves towards the next closest pixel employing the fast marching method (FMM)33. cv.INPAINT TELEA function of cv2 library is used for inpainting. Figure 3 illustrates the process of hair removal using DHR 34.
Data Augmentation
A huge challenge with dermoscopic datasets is the small number of labeled lesion images. The main aim of augmentation is to obtain an optimal performance with deep learning, which requires huge number of training lesion images.To overcome the issue of data limitation and class skewness in the data, augmentation isperformed on the images after the hair-removal process. Applying the image augmentation on the dataset doesnot alter the semantic meaning of the lesion images. The scale and the position of a skin lesion within an image maintains its semantic meaning and doesnot effect classification35. Thus, the dataset is enlarged by transforming the original images towards generating new images with similar labels of actual one by applyingrotation, vertical-horizontal flips, horizontal-vertical shear in the training set and testing set for each class of ISIC-16 and PH2 dataset. The total number of images before and after augmenting the images are listed in Table 1.
Table 1: Dataset Augmentation of PH2 and ISIC-16
| Dataset | Classes | Actual Class ratio | Class ratio after
augmentation |
Train-set
(80%) |
Test-set
(20%) |
Total images before augmentation | Total images after augmentation |
| PH2 | AN | 80 | 2880 | 2304 | 576 | 200 | 8640 |
| CN | 80 | 2880 | 2304 | 576 | |||
| Mel | 40 | 2880 | 2304 | 576 | |||
| ISIC-16 | Be | 1022 | 4009 | 3207 | 802 | 1271 | 8018 |
| Mel | 249 | 4009 | 3207 | 802 |
Segmentation using the Watershed method
Watershed 36is a region-based method which uses image-morphology. The method comprises of following steps:
Gradient magnitude of input image
It finds pixel boundaries. Gradient represents high pixel-value while the pixel direction is represented by the Magnitude.
Dilation and Erosion
Several morphological operations can be performed using dilation and erosion like opening, closing and decomposing of the shape.
Erosion
It removes the minute infected region for preprocessing the image with a highly infected area.
Dilation and Complement
It enlarges the size of the object and fills small holes and narrow gulfs in objects.
Figure 4 shows the sample before and after segmentation images in both the datasets.
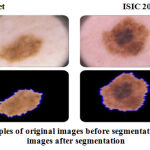 |
Figure 4: (a) and (b) Samples of original images before segmentation, (c) and (d) Samples of images after segmentation. |
Feature Extraction using GLCM for Skin Lesion Detection
The gray-level co-occurrence matrix (GLCM)37, or gray-level spatial dependence matrix (GLDM) is a statistical method used for analyzing the texture of the image. The GLCM is a computation of how many times the distinct combinations of grey level values occurred in an image.Once the GLCM is created, numerous statistical measures can be derived from the GLCM matrix, which gives details regarding the texture of images. Table 2 presentsthe features calculated from the GLCM in this experiment:
 |
Table 2: GLCM features. |
Classification Methods
The following are the architectures used in performing the experiment.
Machine Learning Models
Support Vector Machine (SVM)
Support Vector Machine (SVM) is a binary hyperplane classifier. Given a labeled dataset, SVM will generate an optimal hyperplane to classify the data into certain classes. In order to classify nonlinear data, the data is transformed into a linear form; these transformation functions are called SVM kernels 3839. The SVM relies on structural risk minimization whose goal is to search for a classifier that minimizes the boundary of the expected error 40. In this research, the penalty parameter of SVM is set to C =1.0, using a linear kernel used with the degree=3, gamma =1, and random state=0.
Random Forest (RF)
Random Forest is another machine learning classifier used for classification. It is a classifier that contains multiple decision trees where the number of decision trees is fixed. The decision trees choose an optimal attribute to maximize the Information gain at each level 41. The purity of the dataset is maximized. Random forest is considered as a variant of the Bagging that is used for the formation of decision trees 3942. In this experiment, the splitting criterion is set to ‘gini’ with max_depth and random_state = ‘none’ and n_estimators =’auto’.
k-NN
K-nearest neighbors (KNN) 4318 is one of the simplest Machine Learning algorithms. KNN is known for its late learning methodology. KNN has typically been used in literature for both classification and regression. Typical implementation considers the distance of data points in a scatter plot as a matric to predict the class label in case of classification or the output value inregression. Some of the crucial parameters that must be considered before implementing any such algorithm are the distance threshold, and value of K as they significantly impact the results of models 44. In this research, the value of parameter k, which indicates the number of nearest neighbors, is set to n_neighbors=3 using Euclidean distance p=2.
Logistic Regression (LR)
Logistic Regression (LR)45 is another old stochastic classification algorithm 46, which uses a combination of independent variables as features to perform classification. Unlike Linear regression which predicts the value in a larger range, Logistic regression typically predicts the probability of an outcome in (0,1) range and works on the categorical outcome47484945. Likewise, in SVM, in LR the penalty parameter is set as C=1.0 with solver=’liblinear’ andrandom_state =0.
Deep Learning-based Transfer Learning Models
CNN
Convolutional neural networks 50, or CNNs51, are grid ANN for processing the data with a known grid-like topology. Convolutional networks combine local receptive fields, parameter sharing along with spatial or temporal subsampling 52. The convolution operation is typically denoted with an asterisk:
s(t)=(X*W)(t) (5)
LeNet-5
LeNet 5354 architecture was used primarily. The input size of each image is120 X 160-pixel. The pixel indents are normalized with concerning 255,the black color pixel is associated with a pixel value of 0, whereas a white color pixel is associated with a pixel value of 1. LeNet consists of 3 blocks of convolution, maxpool with Relu5556 activation function.
VGG 16
VGG-16 architecture is chosen as a classifier in this research work as it gives better generalization with other datasets. The network’s input layer needs an RGB image of 224*224 pixels. The source image, which is an input image,is undergone through five convolutional blocks. Small convolutional filters require a filter size of 3*3. A variation occurs between severalfilters’ midst blocks. ReLU (Rectified Linear Unit) is fitted with all the hidden layers, which act as an activation function layer and include spatial pooling. The network ends up with a classifier block which comprises three fully connected (FC) layers 57.
VGG 19
VGG-16 and VGG-19 both are analogous to each other as they share common properties. VGG-19 differs from VGG-16 in terms of a greater number of ConvNet layers, making it work faster comparatively VGG-16. The more thenumber of layers, the lower the learning rate and the loss function. A remarkable amount of training data is required to restrain the effects like over-fitting (as a sequel of numerous free parameters), which introduces the needfor larger training time. Transfer learning overcomes this problem by performing a fine-tune training of pretrained networks with a target database 5859.
Inception V3
Inception is a method thatis used in LeNet architecture. Inception is a convolutional neural network thatrecognizes the framework in images. Serving as a multi-level feature extractor is the main aim of the inception module. The aim is fulfilled by computing 11, 33, and 55 convolutions within the same network segment. The heaped results from these filters are then fed into another layer in the network. Inception gains its strength by associating the devlopment of several convolutional networks of different size in the inception block 60.
Exception
Extreme Inception can be another name for Exception architecture, following the concept of inception module.This architecture is comprising of 36 convolutional layers. For experimentation, Logistic Regression is used for classification. These 36 convolutional layers are then grouped into 14 segments, which are connected except for the first and last segments. Exception architecture is an architecture of the stack of divisible CNN layers with residual connections. This functionality makes the architecture easy to understand and can be modified easily 61
Performance Evaluation Measures
Several measures are used in this experiment for evaluating the performance of the employed classifiers including accuracy, sensitivity, specificity, Dice coefficient, and Jacquard Index62. Eq 1-5 defines the metric for evaluating results. These metrics are acquired from the confusion matrix shown in Table 3, where stands for True Positive, True Negative, False Positive, and False Negative respectively.
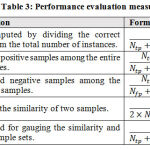 |
Table 3: Performance evaluation measures. |
Experimental Results
This section discussed the performance of the designed CAD-MD system and compared it with several state-of-the-art classification methods on the benchmark publicly available ISIC 2016 and PH2 datasets. The experiments were performed using Python 3.2 with Jupyter Notebook using the libraries namely Keras, Sklearn (scikit-learn), cv2 (OpenCV), Scipy, os, Random, Matplotlib.The models are trained on a GPU comprising given specification- Quadro P4000 NVidia 14 core GPU with 8 GB graphics memory, Intel Xeon Dual Processor workstation 2.5 GHz, DDR4-RAM 64 GBwith Windows 10 Home edition.CUDA and cuDNNare the Python wrappers needed for programming with Nvidia GPU’s. The models are designed using the Keras Library with the Tensorflow framework. Every model is trained to normalize the gradients using the Rmsprop optimizer, and a suitable dropout 0.5 is used to reducethe overfitting problem in Transfer learning.
Dataset details
The ISIC 2016 and PH2 benchmark publicly available datasets are utilized for experimentation in this work. The ISIC and PH2 dataset comprise 1271 and 200 actual RGB images respectively. The class ratio discussed in Table 1 represents skewness in both the datasets,comprising benign lesion images in majority. Therefore, to rebalance the dataset, images are augmented using the operations namely-rotation, vertical – horizontal flips, and horizontal vertical shear operations. After the augmentation, the ISIC and PH2 training dataset comprises 8018, 8640 images respectively. In ISIC dataset, 1604 images are used for testing which is 20% of the dataset and 6414 images are used for training which is 80% of the dataset. Similarly, in PH2 dataset, 1728 images are used for testing which is 20% of the dataset and 6912 images are used for testing which is 80% of the dataset. For validation, 5fold cross-validation is used.
Parameter Setting
Table 4 shows the parameters tuned for modeling the machine learning classifier’sbehavior and deep learning-based transfer learning models for effective and accurate classification.
Table 4: Tuned parameters forthe machine learning and deep learning-based transfer learning models.
| Learning Paradigm | Classifiers | Parameters |
| Machine Learning Models | RF | bootstrap=True, criterion=’gini’, max_depth=2, n_estimators=10, random_state = 0 |
| LR | penalty = ‘l2’, C = 1, random_state = 0 | |
| K-NN | n_neighbors=5, p=2 | |
| SVM | C=1, kernel=’linear’, degree=3, gamma=1, random_state = 0 | |
| Deep Learning-Based Transfer Learning Models | LeNet-5 | Optimizing function- RMSProp, learning rate-0.001, batch size-32, Dropout 0.5, loss function- Categorical Cross entropy, epochs- 200 |
| VGG-16 | Optimizing function- SGD, learning rate-0.001, batch size-32, Dropout 0.7, loss function- Categorical Cross entropy, epochs- 200 | |
| VGG-19 | Optimizing function- RMSProp, learning rate-0.001, batch size-32, Dropout 0.7, loss function- Categorical Cross entropy, epochs- 250 | |
| Inception V3 | Optimizing function- SGD, learning rate-0.0001, batch size-16, Dropout 0.5, loss function- Categorical Cross entropy, epochs- 350 | |
| Exception | Optimizing function- SGD, learning rate-0.0001, batch size-16, Dropout 0.5, loss function- Categorical Cross entropy, epochs- 300 |
Classification result using machine learning and deep learning-based transfer leaning modelson PH2 and ISIC 16 datasets
Two set of experimentations are carried out in this workon PH2 and ISIC-2016 using machine learning and deep learning-based transfer leaning models.For the classification of pigmented skin lesions, four state-of-the-art machine learning classifiers namely, RF, LR, K-NN, and SVM, and deep learning-based transfer learning models namely Inception, Xception, VGG-16, VGG-19 and LeNet-5 are used. The first set of experiment is conducted on PH2 dataset with augmented and without-augmented dataset using machine and deep learning-based transfer leaning models.Similarly, the second set of experimentation is performed on ISIC-2016 datasetusing machine and deep learning-based transfer leaning models on augmented and actual dataset.
PH2 Dataset
In first set of experiment,we evaluated the designed CAD-MD system’s efficiencyusing machine learning and deep learning-based transfer learning models on PH2 dataset on actual and augmented images. Results of with and without augmented PH2 dataset using machine learning and deep learning-based transfer learning models is reported in Table-5. Results obtained using machine learning and deep learning models on actual images are comparatively poor than the results obtained using augmented dataset.In machine learning models, Logistic Regression has outperformed with an accuracy of 62.5% on actual dataset, whilek-NN surpasseswith an accuracy of 66.7% on augmented dataset.Likewise, in deep learning-based transfer leaning models, VGG16 has outperformed with an accuracy of 95.7% on actual dataset, and performed superior on augmented dataset with an accuracy of 99.1%. The performance of machine learning and deep learning-based transfer learning models on actual and augmented PH2 dataset is graphically shown in Figure 5 and Figure 6 respectively.
Table 5: Results of with and without augmented PH2 dataset using machine learning and deep learning-based transfer learning models
|
PH2 Dataset |
||||||||||
| Dataset | Measures | Machine Learning Models | Deep Learning based Transfer Learning Models | |||||||
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | ||
| Actual Dataset | ACC | 53.7 | 62.5 | 46.6 | 61.2 | 90.4 | 92.5 | 91.4 | 95.7 | 55.2 |
| SE | 48.5 | 50.2 | 45.7 | 54.8 | 91.2 | 89.4 | 90.5 | 89.2 | 49.9 | |
| SP | 63.9 | 69.9 | 59.3 | 77.5 | 93.5 | 93.1 | 95.1 | 97.1 | 70.2 | |
| DI | 34.6 | 42.3 | 33.5 | 46.4 | 65.6 | 67.1 | 64.6 | 68.1 | 38.3 | |
| JA | 26.9 | 29.6 | 23.8 | 33.3 | 54.4 | 59.1 | 54.9 | 60.2 | 27.2 | |
|
Augmented Dataset |
ACC | 58.3 | 62.5 | 50.0 | 66.6 | 98.4 | 98.9 | 98.0 | 99.1 | 60.4 |
| SE | 53.1 | 54.8 | 49.6 | 59.4 | 95.3 | 93.5 | 94.6 | 94.3 | 55.2 | |
| SP | 68.5 | 66.4 | 60.1 | 79.5 | 98.8 | 98.4 | 97.5 | 99.5 | 73.5 | |
| DI | 40.3 | 45.1 | 38.7 | 49.6 | 68.8 | 70.3 | 68.1 | 71.6 | 42.5 | |
| JA | 31.5 | 34.2 | 28.4 | 39.6 | 60.7 | 64.3 | 60.1 | 65.4 | 32.3 | |
|
Note: C1=SVM, C2= LR, C3 = RF, C4 = KNN, C5 = Xception, C6 = Inception, C7 = VGG19, C8 = VGG16, C9 = LeNet-5 |
||||||||||
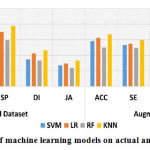 |
Figure 5: Performance of machine learning models on actual and augmented PH2 dataset. |
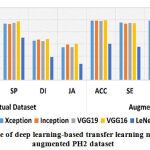 |
Figure 6: Performance of deep learning-based transfer learning models on actual and augmented PH2 dataset. |
ISIC16 Dataset
The designed CAD-MD system’s efficiency using machine learning and deep learning-based transfer learning models on ISIC 16 dataset on actual and augmented images is carried out in second set of experiment. Results of with and without augmented ISIC 16 dataset using machine learning and deep learning-based transfer learning models is reported in Table-6. Likewise, theresults reported in Table 5, results demonstrated in Table 6 shows poor performance obtained on actual images than the results obtained using augmented dataset. In machine learning models, k-NN outperformed superior on both without-augmented and augmented dataset with an accuracy of 56.7% and 61.6% respectively. WhileLeNet-5 has outperformed with an accuracy of 77.9% on actual dataset, and performed superior on augmented dataset with an accuracy of 82.5%. The performance of machine learning and deep learning-based transfer learning models on actual and augmented ISIC 16 dataset is graphically shown in Figure 7 and Figure 8 respectively.
Table 6: Results of with and without augmented ISIC 16 dataset using machine learning and deep learning-based transfer learning models.
|
ISIC 2016 Dataset |
||||||||||
| Dataset | Measures | Machine Learning Models | Deep Learning based Transfer Learning Models | |||||||
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | ||
| Actual Dataset | Acc | 47.6 | 45.4 | 52.4 | 56.7 | 62.9 | 63.2 | 61.5 | 63.5 | 77.9 |
| SE | 46.8 | 46.8 | 47.5 | 55.7 | 57.1 | 58.7 | 56.3 | 60.1 | 73.1 | |
| SP | 60.7 | 62.6 | 67.6 | 68.9 | 67.2 | 68.8 | 69.3 | 74.1 | 85.3 | |
| DI | 33.4 | 32.9 | 41.3 | 43.1 | 33.1 | 34.7 | 31.6 | 36.8 | 55.6 | |
| JA | 28.9 | 27.8 | 30.4 | 29.1 | 27.1 | 29.8 | 27.4 | 32.2 | 50.3 | |
|
Augment Dataset |
Acc | 53.3 | 50.0 | 56.6 | 61.6 | 67.5 | 67.8 | 66.6 | 68.1 | 82.5 |
| SE | 52.4 | 51.4 | 52.6 | 60.3 | 61.2 | 62.8 | 60.4 | 65.2 | 78.4 | |
| SP | 65.3 | 64.5 | 71.5 | 74.2 | 72.5 | 74.1 | 71.7 | 76.5 | 88.6 | |
| DI | 40.7 | 38.6 | 45.2 | 46.3 | 36.3 | 37.9 | 35.5 | 40.3 | 59.8 | |
| JA | 33.5 | 32.4 | 34.6 | 35.4 | 33.4 | 35.0 | 32.6 | 37.4 | 55.4 | |
|
Note: C1=SVM, C2= LR, C3 = RF, C4 = KNN, C5 = Xception, C6 = Inception, C7 = VGG19, C8 = VGG16, C9 = LeNet-5 |
||||||||||
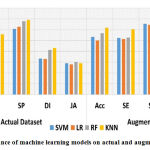 |
Figure 7: Performance of machine learning models on actual and augmented ISIC16 dataset. |
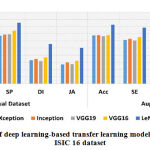 |
Figure 8: Performance of deep learning-based transfer learning models on actual and augmented ISIC 16 dataset. |
Table 5 and Table 6 shows that the use of deep learning-based transfer learning models has improved the accuracy of the proposed model because using transfer learning, we incorporate pre-trained weights that are used for training of every model in deep learning.The performance of all the machine learning and deep learning-based transfer learning architectures is evaluated based on the Accuracy, Sensitivity, Specificity, Jaccard index, and Dice Coefficient.Since VGG16 has achieved the highest promising accuracy than all employed classifiers, we have evaluated receiver operating characteristics (ROC) and area under the ROC curve (AUC-ROC) curve on PH2 data set. Figure 9 demonstrates the ROC curve and AUC-ROC acquired by employing the VGG-16 model with an AUC value of 99.1% on PH2 data set.
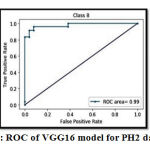 |
Figure 9: ROC of VGG16 model for PH2 dataset. |
Discussion
The designed CAD-MD system have a vital contribution in the early and accurate diagnosis of melanoma for numerous reasons. First,our study comprises of all the vital phases required in the development of an effective automated diagnostic system for the detection and classification of melanoma as benign or malignant. Second, it provides useful insights that assists researchers in investigating the importance of data augmentation towards achieving optimal classification performance. Third, our designed system incorporates the potentials of machine learning and deep learning-based transfer learning models in the early and accurate melanoma detection which has been overlooked by the previous studies. CAD-MD aims to provide reliable melanoma diagnosis but also assists the dermatologists and medical experts by providing valuable insights in identifying the melanoma accurately. Since the deep learning-based transfer learning models outperforms the machine learning classifiers, we performed an analysis comparing our proposed work (outperformed model) with previously conducted studies on PH2 and ISIC-2016 datasets, shown in Table 7.
Table 7: Comparison analysis of proposed study with the current study on PH2 and ISIC 16 dataset.
| PH2 | ISIC-16 | |||
| Previous Work | Methods | Accuracy (%) | Methods | Accuracy (%) |
| Inception v4 63 | 98.8 | Inception v4 64 | 73.4 | |
| VGG19 65 | 90.5 | VGG19 66 | 76.1 | |
| VGG 16 67 | 93.1 | VGG16 68 | 79.2 | |
| LeNet-5 69 | 75.2 | VGG16 57 | 81.3 | |
| Proposed Work | VGG 16 | 99.1 | LeNet-5 | 82.5 |
Conclusion and Future Work
In this work an effective CAD-MD system is designed towards improving the classification of melanoma and exploring the influence of data augmentation on the performance of four machine learning models namely SVM, Logistic Regression, KNN, and Random Forest and five deep learning-based transfer learning models namely LeNet-5, VGG-16, VGG-19, Inception and Xception.Our designed work overcomes the data limitation and skewness issue by incorporating data augmentation. Two set of experimentations are conducted on PH2 and ISIC16 datasets to evaluate the effectiveness of all employed classifiers with and without image augmentation. Empirical results prove that enhanced classification has been achieved with augmented images, where, deep learning-based transfer learning models outperforms machine learning classifiers. The CNN VGG16 modeloutperforms on PH2 dataset with an accuracy of 99.13%. The performance of classifiersis verified using appropriate measures namely accuracy, sensitivity, specificity, jacquard index, and dice coefficient.The limitation of our work is that towards designing CAD-MD system we have reviewed the work published tillAugust 2020. Since, the CAD system is a rapid emerging field of technology, our work has missed some recent work conducted after August 2020 to April 2021 in the field of melanoma detection and classification. Future work can be accommodated using new deep learning models for transfer learning to improve the results on different datasets of medical diseases. In future, the designed CAD system can further be enhanced for smart devices. The designed CAD system can also be used to diagnose several other severe skin lesion types, which vary in different skin colors.
Conflict of Interest
The Authors declares that they have no conflict of interest.
Funding source
There are no funding Source for this article.
References
- Mendonca T, Ferreira PM, Marques JS, Marcal ARS, Rozeira J. PH2 – a dermoscopic image database for research and benchmarking. In: Conference Proceedings : … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Vol 2013. ; 2013:5437-5440. doi:10.1109/EMBC.2013.6610779
CrossRef - Li Y, Shen L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors. 2018;18(2):556.
CrossRef - Mishra R, Daescu O. Deep learning for skin lesion segmentation. In: Bioinformatics and Biomedicine (BIBM), 2017 IEEE International Conference On. ; 2017:1189-1194.
- Amelard R, Glaister J, Wong A, Clausi DA. High-Level Intuitive Features (HLIFs) for intuitive skin lesion description. IEEE Transactions on Biomedical Engineering. 2015;62(3):820-831. doi:10.1109/TBME.2014.2365518
CrossRef - Noar SM, Leas E, Althouse BM, Dredze M, Kelley D, Ayers JW. Can a selfie promote public engagement with skin cancer? Preventive Medicine. 2018;111(November):280-283. doi:10.1016/j.ypmed.2017.10.038
CrossRef - Bhattacharya A, Young A, Wong A, Stalling S, Wei M, Hadley D. Precision diagnosis of melanoma and other skin lesions from digital images. In: AMIA Summits on Translational Science Proceedings. Vol 2017. American Medical Informatics Association; 2017:220.
- Olga Russakovsky, Jia Deng, Hao Su, Jonathan Krause, Sanjeev Satheesh, Sean Ma, Zhiheng Huang AK, Aditya Khosla, Michael Bernstein, Alexander C. Berg LF-F. ImageNet Large Scale Visual Recognition Challenge. International Journal of Computer Vision. 2015;115(3):211-252. doi:10.1007/s11263-015-0816-y
CrossRef - Fujisawa Y, Inoue S, Nakamura Y. The Possibility of Deep Skin Tumor Classifiers. Frontiers in Medicine. 2019;6(August):1-10. doi:10.3389/fmed.2019.00191
CrossRef - Moi Hoon Yap, Gerard Pons, Joan Marti, Sergi Ganau, Melcior Sentis, Reyer Zwiggelaar, Adrian K. Davison RM. Automated Breast Ultrasound Lesions Detection Using Convolutional Neural Networks. IEEE Journal of Biomedical and Health Informatics. 2018;22(4):1218-1226. doi:10.1109/JBHI.2017.2731873
CrossRef - Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. In: European Conference on Computer Vision. ; 2014:818-833.
- Cheng PM, Malhi HS. Transfer learning with convolutional neural networks for classification of abdominal ultrasound images. Journal of digital imaging. 2017;30(2):234-243.
CrossRef - Y. Fujisawa, Y. Otomo, Y. Ogata, Y. Nakamura, R. Fujita, Y. Ishitsuka, R. Watanabe, N. Okiyama, K. Ohara M, Fujimoto. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. British Journal of Dermatology. 2019;180(2):373-381.
CrossRef - Andre Esteva, Brett Kuprel, Roberto A. Novoa, Justin Ko, Susan M. Swetter, Helen M. Blau ST. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115-118. doi:10.1038/nature21056
CrossRef - Baltruschat IM, Nickisch H, Grass M, Knopp T, Saalbach A. Comparison of deep learning approaches for multi-label chest X-ray classification. Scientific reports. 2019;9(1):1-10.
CrossRef - Abuzaghleh O, Barkana BD, Faezipour M. Automated skin lesion analysis based on color and shape geometry feature set for melanoma early detection and prevention. 2014;(May). doi:10.1109/LISAT.2014.6845199
CrossRef - Ballerini L, Fisher RB, Aldridge B, Rees J. A Color and Texture Based Hierarchical K-NN Approach to the Classification of Non-melanoma Skin Lesions. Lecture Notes in Computational Vision and Biomechanics. 2013;6:63-86. doi:10.1007/978-94-007-5389-1_4
CrossRef - Zhang, Jianpeng and Xie, Yutong and Wu, Qi and Xia Y. Skin lesion classification in dermoscopy images using synergic deep learning. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. ; 2018:12-20.
- Sumithra R, Suhil M, Guru DS. Segmentation and classification of skin lesions for disease diagnosis. Procedia Computer Science. 2015;45(C):76-85. doi:10.1016/j.procs.2015.03.090
CrossRef - Almaraz-Damian J-A, Ponomaryov V, Sadovnychiy S, Castillejos-Fernandez H. Melanoma and nevus skin lesion classification using handcraft and deep learning feature fusion via mutual information measures. Entropy. 2020;22(4):484.
CrossRef - Hosny KM, Kassem MA, Foaud MM. Skin Cancer Classification using Deep Learning and Transfer Learning. In: 2018 9th Cairo International Biomedical Engineering Conference (CIBEC). ; 2018:90-93.
- Ray A, Gupta A, Al A. Skin Lesion Classification With Deep Convolutional Neural Network: Process Development and Validation. JMIR Dermatology. 2020;3(1):e18438.
- Zunair H, Hamza A Ben. Melanoma detection using adversarial training and deep transfer learning. Physics in Medicine & Biology. 2020;65(13):135005.
CrossRef - Sagar A. Convolutional Neural Networks for Classifying Melanoma Images. bioRxiv. Published online 2020:1-12.
- Ratul AR, Mozaffari MH, Lee W-S, Parimbelli E. Skin Lesions Classification Using Deep Learning Based on Dilated Convolution. bioRxiv. Published online 2019:860700.
- Guo G, Razmjooy N. A new interval differential equation for edge detection and determining breast cancer regions in mammography images. Systems Science & Control Engineering. 2019;7(1):346-356.
CrossRef - Navid Razmjooy, Mohsen Ashourian, Maryam Karimifard, Vania V Estrela, Hermes J Loschi, Douglas do Nascimento, Reinaldo P França MV. Computer-Aided Diagnosis of Skin Cancer: A Review. Current Medical Imaging. 2020;16(7):781-793. doi:DOI: 10.2174/1573405616666200129095242
CrossRef - Rashid Sheykhahmad F, Razmjooy N, Ramezani M. A novel method for skin lesion segmentation. International Journal of Information, Security and Systems Management. 2015;4(2):458-466.
- Razmjooy N, Sheykhahmad FR, Ghadimi N. A hybrid neural network–world cup optimization algorithm for melanoma detection. Open Medicine. 2018;13(1):9-16.
CrossRef - Razmjooy N, Mousavi BS, Soleymani F, Khotbesara MH. A computer-aided diagnosis system for malignant melanomas. Neural Computing and Applications. 2013;23(7-8):2059-2071.
- Fotouhi S, Asadi S, Kattan MW. A comprehensive data level analysis for cancer diagnosis on imbalanced data. Journal of biomedical informatics. 2019;90:103089.
CrossRef - Mojdeh Rastgoo, Guillaume Lemaitre JM, Morel O, Marzani F, Rafael, Garcia, Meriaudeau F. Tackling the problem of data imbalancing for melanoma classification. In: BIOSTEC – 3rd International Conference on BIOIMAGING. ; 2016:1-9.
- David Gutman, Noel C. F. Codella, Emre Celebi, Brian Helba, Michael Marchetti, Nabin Mishra AH. Skin Lesion Analysis toward Melanoma Detection: A Challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC). arXiv preprint arXiv:160501397. 2016;April:168-172. doi:10.1109/ISBI.2018.8363547
CrossRef - Telea A. An Image Inpainting Technique Based on the Fast Marching Method. Journal of Graphics Tools. 2004;9(1):23-34. doi:10.1080/10867651.2004.10487596
CrossRef - Schmid-Saugeona P, Guillodb J, Thirana, J-P. Towards a computer-aided diagnosis system for pigmented skin lesions. Computerized Medical Imaging and Graphics. 2003;27(1):65-78. doi:10.1016/S0895-6111(02)00048-4
CrossRef - Tri-Cong Pham, Chi-Mai Luong, Muriel Visani V-DH. Deep CNN and Data Augmentation for Skin Lesion Classification. In: Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). ; 2018:573-582. doi:10.1007/978-3-319-75420-8_54
CrossRef - Lou, Shan and Pagani, Luca and Zeng, Wenhan and Jiang, X and Scott P. Watershed segmentation of topographical features on freeform surfaces and its application to additively manufactured surfaces. Precision Engineering. 2020;63:177-186.
CrossRef - Singh S, Srivastava D, Agarwal S. GLCM and its application in pattern recognition. In: 5th International Symposium on Computational and Business Intelligence, ISCBI. ; 2017:20-25. doi:10.1109/ISCBI.2017.8053537
CrossRef - Codella, Noel and Cai, Junjie and Abedini, Mani and Garnavi, Rahil and Halpern, Alan and Smith JR. Deep Learning, Sparse Coding, and SVM for Melanoma Recognition in Dermoscopy Images. In: International Workshop on Machine Learning in Medical Imaging. ; 2015:118-126. doi:10.1007/978-3-319-24888-2
CrossRef - Oliveira RB, Papa JP, Pereira AS, Tavares JMRS. Computational methods for pigmented skin lesion classification in images: review and future trends. Neural Computing and Applications. 2018;29(3):613-636. doi:10.1007/s00521-016-2482-6
CrossRef - O. A, B.D. B, M. F. Noninvasive real-time automated skin lesion analysis system for melanoma early detection and prevention. IEEE Journal of Translational Engineering in Health and Medicine. 2015;3(March). doi:10.1109/JTEHM.2015.2419612
CrossRef - Xia J, Ghamisi P, Yokoya N, Iwasaki A. Random forest ensembles and extended multiextinction profiles for hyperspectral image classification. IEEE Transactions on Geoscience and Remote Sensing. 2018;56(1):202-216. doi:10.1109/TGRS.2017.2744662
CrossRef - Mahbod A, Schaefer G, Wang C, Ecker R, Ellinge I. Skin Lesion Classification Using Hybrid Deep Neural Networks. In: IEEE International Conference on Acoustics, Speech and Signal Processing, ICASSP. ; 2019:1229-1233. doi:10.1109/ ICASSP.2019. 8683352
CrossRef - Peruch F, Bogo F, Bonazza M, Cappelleri VM, Peserico E. Simpler, faster, more accurate melanocytic lesion segmentation through MEDS. IEEE Transactions on Biomedical Engineering. 2014;61(2):557-565. doi:10.1109/TBME.2013.2283803
CrossRef - Pathan S, Prabhu KG, Siddalingaswamy PC. Techniques and algorithms for computer aided diagnosis of pigmented skin lesions—A review. Biomedical Signal Processing and Control. 2018;39(August):237-262. doi:10.1016/j.bspc.2017.07.010
CrossRef - Press, S James and Wilson S. Choosing between Logistic Regression and Discriminant Analysis. Journal of the American Statistical Association. 1978;73(364):699-705.
CrossRef - Baştanlar Y, Özuysal M. Introduction to machine learning. MicroRNA Biology and Computational Analysis Methods in Molecular Biology. 2013;1107:105-128. doi:10.1007/978-1-62703-748-8_7
CrossRef - Dreiseitl S, Ohno-Machado L, Kittler H, Vinterbo S, Billhardt H, Binder M. A comparison of machine learning methods for the diagnosis of pigmented skin lesions. Journal of Biomedical Informatics. 2001;34(1):28-36. doi:10.1006/jbin.2001.1004
CrossRef - Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: A methodology review. Journal of Biomedical Informatics. 2002;35(5-6):352-359. doi:10.1016/S1532-0464(03)00034-0
CrossRef - Kurt I, Ture M, Kurum AT. Comparing performances of logistic regression, classification and regression tree, and neural networks for predicting coronary artery disease. Expert Systems with Applications. 2008;34(1):366-374. doi:10.1016/j.eswa.2006.09.004
CrossRef - Hoo-Chang Shin, Holger R. Roth, Mingchen Gao, Le Lu ZX, Isabella Nogues, Jianhua Yao, Daniel Mollura RMS. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE transactions on medical imaging. 2016;35(5):1285-1298.
CrossRef - Jafari, MH and Nasr-Esfahani, E and Karimi, N and Soroushmehr, SMR and Samavi, S and Najarian K. Extraction of Skin Lesions from Non-Dermoscopic Images Using Deep Learning. arXiv preprint arXiv:160902374. Published online 2016:10-11. doi:10.1007/s11548-017-1567-8
CrossRef - Gao M, Xu Z, Lu L, Harrison AP, Summers RM, Mollura DJ. Deep Learning, Sparse Coding, and SVM for Melanoma Recognition in Dermoscopy Images. 2017;(October). doi:10.1007/978-3-319-24888-2
CrossRef - Forest Agostinelli, Matthew Hoffman, Peter Sadowski PB. Learning Activation Functions to Improve Deep Neural Networks. In: International Conference on Learning Representations (ICLR). ; 2014:1-10. doi:10.1007/3-540-49430-8
CrossRef - Yangqing Jia, Evan Shelhamer, Jeff Donahue SK, Jonathan Long, Ross Girshick, Sergio Guadarrama TD. Caffe: Convolutional Architecture for Fast Feature Embedding. In: Proceedings of the 22nd ACM International Conference on Multimedia. ; 2014:675-678. doi:10.1145/2647868.2654889
CrossRef - Krizhevsky A, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. NIPS’12 Proceedings of the 25th International Conference. 2012;1:1-9.
- Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size. Published online 2016:1-13. doi:10.1007/978-3-319-24553-9
CrossRef - Romero-Lopez A, Giro-i-Nieto X, Burdick J, Marques O. Skin Lesion Classification from Dermoscopic Images Using Deep Learning Techniques. Proceedings of the lASTED International Conference Biomedical Engineering (BioMed 201 7) February 20 – 21, 2017. Published online 2017:49-54. doi:10.2316/P.2017.852-053
CrossRef - Sun Y, Wang X, Tang X. Deep Learning Face Representation From Predicting 10 000 Classes. Cvpr. Published online 2014:1891-1898. doi:10.1109/CVPR.2014.244
CrossRef - Galea C, Farrugia RA. Matching Software-Generated Sketches to Face Photographs with a Very Deep CNN, Morphed Faces, and Transfer Learning. IEEE Transactions on Information Forensics and Security. 2018;13(6):1421-1431. doi:10.1109/TIFS.2017 .2788002
CrossRef - Alom MZ, Hasan M, Yakopcic C, Taha TM. Inception Recurrent Convolutional Neural Network for Object Recognition. Published online 2017. http://arxiv.org/abs/1704.07709.
- Chollet F. Xception: Deep learning with depthwise separable convolutions. Proceedings – 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. 2017;2017-Janua:1800-1807. doi:10.1109/CVPR.2017.195
CrossRef - Tharwat A. Classification assessment methods. Applied Computing and Informatics. Published online 2020:1-14. doi:10.1016/j.aci.2018.08.003
CrossRef - Tallha Akram, Hafiz M. Junaid Lodhi, Syed Rameez Naqvi, Sidra Naeem, Majed Alhaisoni MA, Qadri, Sajjad Ali Haider NN. A multilevel features selection framework for skin lesion classification. Human-centric Computing and Information Sciences. 2020;10:1-26.
CrossRef - Pham TC, Luong CM, Visani M, Hoang VD. Deep CNN and Data Augmentation for Skin Lesion Classification. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2018;10752 LNAI(June):573-582. doi:10.1007/978-3-319-75420-8_54
CrossRef - dos Santos FP, Ponti MA. Robust feature spaces from pre-trained deep network layers for skin lesion classification. In: 2018 31st SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI). ; 2018:189-196.
- Goyal M, Knackstedt T, Yan S, Oakley A, Hassanpour S. Artificial Intelligence-Based Image Classification for Diagnosis of Skin Cancer: Challenges and Opportunities. Published online 2019. http://arxiv.org/abs/1911.11872.
- Rodrigues D de A, Ivo RF, Satapathy SC, Wang S, Hemanth J, Rebouças Filho PP. A new approach for classification skin lesion based on transfer learning, deep learning, and IoT system. Pattern Recognition Letters. Published online 2020:1-11.
- Menegola A, Fornaciali M, Pires R, Bittencourt FV, Avila S, Valle E. Knowledge transfer for melanoma screening with deep learning. In: 14th International Symposium on Biomedical Imaging (ISBI ). ; 2017:297-300. doi:10.1109/ISBI.2017.7950523
CrossRef - Nylund A. To be, or not to be Melanoma : Convolutional neural networks in skin lesion classification. Published online 2016.