Yuvarani Thambidurai1, Sudarsanam Durairaj2, Siddhardha Solosan S3, Sewali Ghosh4 and Habeeb Shaik Mohideen5*
1Department of Bioinformatics, Bharathiar University, Coimbatore, Tamilnadu – 641 046, India.
2Department of Advanced Zoology and Biotechnology, School of Genomics and Bioinformatics, Loyola College, Chennai, Tamilnadu – 600 034, India.
3Institute of Microbiology, Madras Medical College, Chennai – 600003 India.
4Department of Advance Zoology and Biotechnology, Guru Nanak College, Velachery, Chennai, Tamilnadu – 600042, India.
5Department of Genetic Engineering, Bioinformatics and Entomoinformatics Lab, SRM Institute of Science and Technology, Kattankulathur, Tamilnadu – 603203, India.
Corresponding Author E-mail: Habeeb_skm@yahoo.co.in
DOI : https://dx.doi.org/10.13005/bpj/2139
Abstract
Identifying new targets and new drugs has always been a daunting task, especially in cancer research. This studyexamines the binding interaction and the drug-likeness properties of small molecules derived from marine sponge Dysideaherbacea to breast cancer receptors: epidermal growth factor receptor and estrogen receptor. The receptor’s interaction with the ligand was evaluated using the Schrodinger Glide package, and affinities were assessed based on the glide score. Ligand molecules that have higher binding affinity were evaluated further for their ADMET properties using Molinspiration.We found multiple ligands binding to these targets; however, Pyridine 3 carboxyamidewas found to have binding affinity to both the receptors. Compared to the other small molecules,further simulation studies could be taken up to ascertain its structural dynamics and ensuing in-vitro experiments that could prove growth inhibition of breast cancer cells.
Keywords
Breast Cancer; Dysideaherbacea; Drug-Likeness; Epidermal Growth Factor Receptor; Estrogen Receptor; Molecular Docking
Download this article as:| Copy the following to cite this article: Thambidurai Y, Durairaj S, Solosan S. S, Ghosh S, Mohideen H. S. Binding Potential of Dysidea Herbacea Derived Small Molecules to Breast Cancer Implicating Targets Estrogen and Epidermal Growth Factor Receptors. Biomed Pharmacol J 2021;14(1). |
| Copy the following to cite this URL: Thambidurai Y, Durairaj S, Solosan S. S, Ghosh S, Mohideen H. S. Binding Potential of Dysidea Herbacea Derived Small Molecules to Breast Cancer Implicating Targets Estrogen and Epidermal Growth Factor Receptors. Biomed Pharmacol J 2021;14(1). Available from: https://bit.ly/39IVmRV |
Introduction
Breast cancer affects women mainly becausespecific cells in the breast tissue behave abnormally due to mutation and multiply rapidly in an uncontrolled manner. It is hormonal cancer where glandular tissues are susceptible to hormonal changes. The expression status of the estrogen receptor (ER) and cell surface tyrosine kinase human epidermal growth factor receptor (EGFR) stays important in employing suitable therapy1.
Estrogens play an essential role in mitogenic activity in both normal and cancerous breast tissues. Numerous studies have reported the potential role of estrogens in the proliferation and progression of breast tumors by provoking multiple growth-promoting signals2,3,4. Studies on estrogen reveal that 17 β-estradiol upregulates c-Myc and cyclinD1 expression and initiates a shift over the cell cycle event from G1 to S phase. Hence, developing drugs based on ER’s structure to inhibit their active process will be worthwhile for treating breast cancer5.
The ER, Progesterone receptor and HER-2 receptor influence in the progression of breast cancer. Among all these receptors, ER serves as a potential target as its expression rate in breast cancer patient is comparatively high.Over expression of ER in 70% breast cancer patients has been observed hence a prime target for developing drugs however, ER fails to exhibit in certain patients6. Under the circumstances, the EGFR plays a significant role in triple negative breast cancer (TNBC). In the absence of triple receptors i.e. TNBC, the level of EGFR is found to be increased which serves as a key target for promising treatment strategies. The EGFR activated by ligand and promotes signal transduction for cellular proliferation, differentiation, migration and its survival. Hence, targeting the ATP binding cleft of the tyrosine kinase domain of EGFR by potential inhibitors led an effective treatment for breast cancer patients7.
The compounds derived from nature especially marine organisms are proven to have potential in treating cancer and serve as better anti-cancer lead molecules8.Molecular docking is a method widely used to study the affinity of molecules on target structure. Considering the role of certain target receptors ER, EGFR in the initiation and progression of breast cancer, small molecules derived from the marine sponge with anticancer properties are examined individually to understand the binding affinity. This study focusses on validating the small molecules reported from our previous study which were derived from the marine sponge, Dysideaherbacea to target the receptors ER and EGFR.
Methodology
Target and ligand structures
Small molecules derived from the marine sponge, Dysideaherbacea,were tested forthe binding affinity with the target receptors. The structures of the small molecules were retrieved from pubchem database in SDF format and were converted in to 3D PDB format. The three-dimensional structure of ER (PDBID-5T97) was downloaded from PDB. However, the structure of EGFR was modelled using I-Tasser9 software by threading method because of lack of experimental structure in PDB database. Both these structures were subjected to energy minimization and were later found to be of acceptable quality based on Ramachandran plot.
Preparation and docking of target and ligand
The target protein was prepared using Schrodinger’s molecular docking software, utilizing the protein preparation wizard. During the target protein preparation, the water molecules and the hetero atoms were removed, charges and bond orders were doled out, hydrogen atoms were added to the stunning particles, thereby achieving exact ionization and tautomeric states of amino acids like serine, glutamic acid, arginine, aspartic acid and histidine to achieve efficient binding 10,11.Similarly, the ligand molecules were prepared using the ligand preparation module of Schrodinger where these ligand molecules were optimized energetically. Molecular docking was performed using Glide module with default parameters and the best poses were selected based on glide score. Importance was also given to the no. of hydrogen bonds formed between the molecules and other parameters as well 12.
Evaluating the small molecule using molinspiration.
The small molecules that had higher binding affinity and docking score with the target receptors were checked using the molinspiration tool for observing its molecular properties, drug likeness and bioactivity. Small molecules such as pyridine 3-carboxamide, 1-Chloro-3-(naphthalen-1-yl) propan-2-one, 13-Oxabicyclo [10.1.0] tridecane were assessed to develop as a drug based on its properties. The canonical SMILES string of each small molecule retrieved from Pubchem database used as an input for Molinspiration to assess the small molecule for drug likeliness properties.
Results
In silico – structures of target receptors in breast cancer.
In the present study, two receptors namely ER and EGFR were chosen as target since ER is active in almost 70% of breast cancer patients and EGFR in TNBC patients which serves as an important target even in the absence of breast cancer receptors such as Estrogen receptor, Progesterone receptor and Her-2 receptor.
Estrogen receptor
In this study, the ER structure was retrieved from PDB database, which stands as a potential target receptor in breast cancer. The ER structure mainly has α-helices (red ribbon) few β-strands (Yellow arrow) and few coils (green wire) represented in Fig. 1A.
Epidermal growth factor receptor
Primary amino acid sequence of EGFR was retrieved from Uniprot database and used as input to I-Tasser server for predicting its secondary structure. The top five models which were generated by I-Tasser were analysed quantitatively by C-score, which is calculated based on the significance of threading template alignments and the convergence parameters of the structure assembly simulations. C-score was in the range of -5, 2, in which higher value usually specifies model with higher confidence. The first model had a better quality with C-Score of -2.95 and C-score >-1.5 specifies a model with good global topology. The predicted EGFR model is shown in Fig. 1B which mainly consists of coils (green wire), few α helices (red ribbons) and β sheets (yellow arrows).
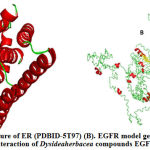 |
Figure 1: (A) Structure of ER (PDBID-5T97) (B). EGFR model generated by I-Tasser Interaction of Dysideaherbacea compounds EGFR |
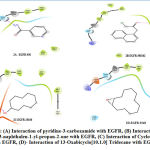
In this study, 3D ligand structures of nine compounds found to be present in D.herbaceawere docked with EGFR and ER using glide software. The compound 1-Chloro-3-naphthalen-1-yl-propan-2-one accepted an h-bond from Met600 to facilitate its interaction (Fig. 2B). Cyclodecanolbinding to EGFR was facilitated by Asp262 that donates hydrogen atom to hydroxyl group of Cyclodecanol (Fig. 2C).The compound 13-Oxabicyclo[10.1.0] tridecane did not show any promising hydrogen bond property though it was able to interact with the receptor in around similar active sites of the target (Fig. 2D).
Similarly, Cholesta-4,6-dien-3-one didn’t form any hydrogen bonding interaction with the active site of target (Fig. 3A). The compound 1,3,4-Triazoleinteracts with the Thr263 of the target (Fig. 3B). Though the compound cyclohexanone had the ability to bind with the receptor, it was only due to non-bonding interactions (Fig. 3C).
Amino acids Asp262 and Thr263 were found to facilitate hydrogen bond formation by donating a hydrogen to Glutaric acid (Fig. 3D). Heptadecanenitrile accepted hydrogen atom from the Thr263 a polar uncharged amino acid in the target receptor (Fig. 3E). The highest binding affinity could be predicted by observing the hydrophobic interactions which are optimised at the cost of hydrogen bonds. On analysing the docking data of the compounds targeted against EGFR, Pyridine-3-Carboxamide and 1-Chloro-3-naphthalen-1-yl-propan-2-one shown better docking and glide score as represented in table 1.
Table 1: Docking data against the target EGFR
| S.No | Compound Name (Pubchem ID) | Docking Score | Ligand Efficiency | Lipo | H bond |
| 1 | Pyridine-3-Carboxamide (936) | -5.376 | -0.597 | -1.089 | -0.32 |
| 2 | 1-Chloro-3-naphthalen-1-yl-propan-2-one (588162) | -5.315 | -0.354 | -1.787 | -0.307 |
| 3 | Cyclodecanol (15166) | -5.14 | -0.467 | -1.535 | -0.18 |
| 4 | 13-Oxabicyclo[10.1.0] tridecane (9248) | -4.815 | -0.37 | -1.719 | 0 |
| 5 | Cholesta-4,6-dien-3-one (3034666) | -4.571 | -0.163 | -2.504 | 0 |
| 6 | 1,3,4-Triazole (20746465) | -3.782 | -0.756 | 0.263 | -0.32 |
| 7 | Cyclohexanone (7967) | -3.617 | -0.517 | -0.407 | 0 |
| 8 | Glutaric acid (743) | -3.109 | -0.345 | -0.488 | -0.299 |
| 9 | Heptadecanenitrile (79388) | 1.742 | 0.097 | -1.852 | -0.118 |
The docking score of Pyridine-3-Carboxamide and 1-Chloro-3-naphthalen-1-yl-propan-2-one were -5.376 and -5.315 respectively, which represented a good binding affinity towards the target receptor. The glide ligand efficiency for Pyridine-3-Carboxamide was -0.597 and 1-Chloro-3-naphthalen-1-yl-propan-2-one was -0.354. The glide lipo for Pyridine-3-Carboxamide was -1.089 and 1-Chloro-3-naphthalen-1-yl-propan-2-one was -1.787. The glide H bond contribution for Pyridine-3-Carboxamide was -0.320 and 1-Chloro-3-naphthalen-1-yl-propan-2-one was -0.307. Data represented in table 1 suggested that the compounds pyridine -3-carboxamide and 1-Chloro-3-naphthalen-1-yl-propan-2-one had better binding affinity as in comparison with the other compounds that were docked against the target.
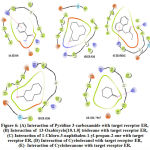
The interaction of pyridine-3-carboxamide ligand with the receptor EGFR represented in Fig. 4 and 5 suggested the ligand bound with the amino acid Val599 by hydrogen bonding at 2.9 Å and in theother instance it bound with Gly598 by forming a hydrogen bonding distance of1.7 Å. Based on the interaction and docking score, it was evident that the ligand pyridine-3-carboxamide has the potential to bind and therefore may inhibit the activity of the target receptor EGFR that aids to possibly treat the breast cancer.
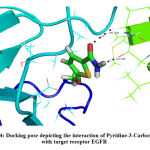 |
Figure 4: Docking pose depicting the interaction of Pyridine-3-Carboxamide with target receptor EGFR. |
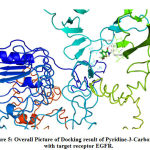 |
Figure 5: Overall Picture of Docking result of Pyridine-3-Carboxamide with target receptor EGFR. |
Interaction of Dysideaherbacea compounds ER
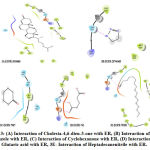
The compound 13-Oxabicyclo[10.1.0] tridecane, it accepts the hydrogen atom from the Arg394 thereby forming hydrogen bond (Fig. 6B). The receptor having Lys449 donated one hydrogen atom to the compound 1-Chloro-3-naphthalen-1-yl-propan-2-one thereby forming hydrogen bond and Arg394 forming pi-cationicinteraction (Fig. 6C). Pro325 facilitates the interaction of ER with Cyclodecanol by forming a h-bond (Fig. 6D) whereas the same was done by Lys449 in the case of Cyclohexanone (Fig. 6E).
The receptor having Lys449 donated one hydrogen atom to the compound 1,3,4-Triazole and glutamic acid accepted one hydrogen atom from the compound (Fig. 7A). In the same way, the compound 26-Nor-5-cholesten-3. beta.-ol-25-one accepts the hydrogen from Lys449 exhibiting a similar interaction (Fig. 7B). Same residues as above Lys449 and Arg394 favoured h-bond with Glutaric acid (Fig. 7C). The receptor exhibiting arginine at the 394th position donated one hydrogen atom to the compound Cholesta-4,6-dien-3-one thereby forming hydrogen bond (Fig. 7D).
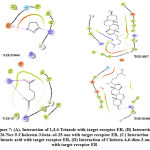
On analysing the docking results of the compounds targeted against ER, Pyridine-3-Carboxamide and 13-Oxabicyclo[10.1.0] tridecane showed better glide score. The docking score of Pyridine-3-Carboxamide and 13-Oxabicyclo[10.1.0] tridecane were -5.372 and -4.934 respectively, which represented a good binding affinity towards the target receptor. The Glide ligand efficiency for Pyridine-3-Carboxamide was -0.597 and 13-Oxabicyclo[10.1.0] tridecane was -0.380. The Glide lipo for Pyridine-3-Carboxamide was -0.592 and 13-Oxabicyclo[10.1.0] tridecane was -1.415. All Data represented in Table 2 suggests the compounds pyridine -3-carboxamide and 13-Oxabicyclo[10.1.0] tridecane had better binding affinity compared with the other compounds that were docked against the target.The interaction of pyridine-3-carboxamide ligand with ER is shown in Fig. 8 and 9 suggested the ligand bound with the receptor at three different amino acids: Lys449 (2.0 Å), Glu353 (1.8 Å) and Arg394 (2.7 Å).
 |
Figure 8: Docking pose of interaction of Pyridine-3-Carboxamide with ER. |
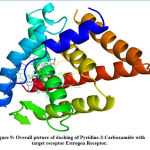 |
Figure 9: Overall picture of docking of Pyridine-3-Carboxamide with target receptor Estrogen Receptor. |
Table 2: Docking data against ER
| S.No | Compound Name (Pubchem ID) | Docking
Score |
Ligand
Efficiency |
Lipo | H bond |
| 1 | Pyridine-3-Carboxamide (936) | -5.372 | -0.597 | -0.592 | -0.379 |
| 2 | 1-Chloro-3-naphthalen-1-yl-propan-2-one (588162) | -4.934 | -0.380 | -1.415 | -0.117 |
| 3 | Cyclodecanol (15166) | -4.746 | -0.316 | -1.073 | -0.160 |
| 4 | 13-Oxabicyclo[10.1.0] tridecane (9248) | -4.450 | -0.405 | 0.467 | -0.320 |
| 5 | Cholesta-4,6-dien-3-one (3034666) | -4.247 | -0.607 | -0.244 | -0.160 |
| 6 | 1,3,4-Triazole (20746465) | -4.063 | -0.813 | -0.405 | -0.160 |
| 7 | Cyclohexanone (7967) | -3.071 | -0.110 | -0.622 | -0.160 |
| 8 | Glutaric acid (743) | -3.067 | -0.341 | -0.524 | -0.361 |
| 9 | Heptadecanenitrile (79388) | -2.154 | -0.077 | -0.433 | 0.000 |
Evaluation of small molecules in molinspiration
The druglikeness of small molecules obtained from marine sponges is assessed for a combination of its molecular properties and structural features. This included hydrophobicity, electronic distribution, hydrogen bonding characteristics, size of the molecule and flexibility; along with other pharmacophoric features influencing the behaviour of a molecule in a living organism that employs bioavailability, transport properties, protein affinity, reactivity, toxicity and metabolic stability.
From table 3 it can be seen that small molecule pyridine 3 carboxamide has miLogP of -0.48, polar surface area of 55.99, molecular weight of 122.13, number of hydrogen bond acceptors is 3, number of hydrogen bond donors is 2, number of molecule violating Lipinski rule is 0, number of rotatable bonds is 1 and molecular volume is 110.16. The bioactivity score of the molecule GPCR ligand is -2.02, Ion channel modulator is -1.51, Kinase inhibitor is -1.70, Nuclear receptor ligand is -2.86, Protease inhibitor is -2.11, and Enzyme inhibitor is -1.42. As the small molecule pyridine 3 carboxamide doesn’t violate Lipinski rule, the molecule is predicted to have druglike properties. Similar properties were witnessed in the case of 1-chloro-3-(naphthalene-1-yl)propan-2-one.
Table 3: Assessing the Drug likeness and bioactive properties of small molecule with high docking score.
| Properties | Pyridine 3
carboxamide |
1-chloro-3-(naphthalene-1-yl)propan-2-one | 13-Oxabicyclo[10.1.0]tridecane |
| miLogP | -0.48 | 3.05 | 4.58 |
| TPSA | 55.99 | 17.07 | 12.53 |
| Natoms | 9 | 15 | 13 |
| MW | 122.13 | 218.68 | 182.31 |
| nON | 3 | 1 | 1 |
| nOHNH | 2 | 0 | 0 |
| Nviolations | 0 | 0 | 0 |
| Nrotb | 1 | 3 | 0 |
| Volume | 110.16 | 194.16 | 201.61 |
| GPCR Ligand | -2.02 | -0.56 | -0.39 |
| Ion channel Modulator | -1.51 | -0.08 | -0.03 |
| Kinase Inhibitor | -1.70 | -0.62 | -0.28 |
| Nuclear receptor Ligand | -2.86 | -0.44 | -0.33 |
| Protease Inhibitor | -2.11 | -0.29 | -0.12 |
| Enzyme Inhibitor | -1.42 | -0.07 | 0.27 |
Discussion
Higher level of estrogen secretion was related with increased risk of breast cancer. Its biological effects includes genesis, progression of malignancy, cell apoptosis and various important roles by binding to ER that occurs in major breast cancer cells 13,14 The ER occurs in two different forms: ER-α and ER-β. Irregular expression of ER-α positive has been reported in about 70% of the breast cancer patients15,16,17,18,19 The ER–α stays as an important receptor in controlling transcription of nuclear DNA for the development of mammary gland19 and it is also an important factor for breast cancer signalling network. Therefore, the inhibition of ER is important for preventing and treating breast cancer20.
The members of estrogen growth family interact specifically with the cell surface receptor EGFR 21. Over expression of EGFR is due to the amplification of EGFR gene that has been involved in breast cancer22. Hence EGFR stands as a prime target for developing active drugs for breast cancer.
Interaction of small molecules with EGFR
Generally, growth factor involves in the regulation of cell division, development and differentiation. The receptors have the capability to identify their specific messenger based on the shape, structure and chemical composition of the active sites. This allows the receptors and the messengers to interact with selectivity. They attach to the active site and inhibit their actions, and block the communication of the message. The binding affinity of each compound was ascertained by observing the glide score and the one which has highest binding affinity may impede the activity of the target receptor23.
On observing the results of docking it was evident that the compound Pyridine-3-Carboxamide accommodating itself around Lys261, Thr263, Cys264 Pro265 and interacts with higher binding affinity with EGFR. The amino radical of pyridine 3-carboxamide donates hydrogen to polar uncharged Thr263 (Fig. 2A).As a result of this interaction, extracellular region could get blocked in autocrine and paracrine growth factor loops, and induce receptor dimerization and downregulation24,25.
The tyrosine kinase domain in EGFR encompasses two lobes that are connected to each other by ten amino acid sequence represented as “hinge”. The NH2 terminal lobe (N-lobe) and COOH terminal is mostly dominated by strands with few helix. In adjacent to the lobes and the hinge, the active site holds the hole-binding domain of ATP, ATP analogues and ATP-competitive inhibitors26. Thus, small molecule tyrosine kinase inhibitors (TKIs) that competitively block ATP binding were designed as potential anticancer agents27,28. As the small molecule pyridine 3 carboxamideimpedes the ATP-binding site of EGFR, it is predicted that it interferes in the downstream signaling process essential for cellular transformation.
Interaction of small molecules with ER
Steroid receptors (SRs), act as ligand-dependent transcription factors, and their activity is associated with the cell cycle29. The ER-α and ER-β are transcription factors that regulate the expression of specific genes in different tissues on a ligand dependent manner30. Several recent studies support the feasibility of using a high throughput screening (HTS) approach to identify small molecules that act directly at the binding interface, or allosterically by inducing a conformational change in the protein. This affects the formation of a functioning macromolecular interface that in-turn impedes ER-α mediated transcription in intact cells. A concentration-dependent inhibition of reporter gene expression upon treatment to sponge extract has been reported31.
The compound Pyridine 3-carboxamideinteracts with ER by accepting hydrogen atom (Lys449) and donating (E353) to ER. Arg394 forms pi cation with the compound which is a noncovalent molecular interaction between the face of the electron rich π systems (Fig. 6A). Hence, pyrimidine 3 carboxamide by binding at the ligand binding site, the estrogen receptor is predicted to be inactivated for further gene expression and thereby the growth and proliferation of breast cancer cells is halted.Based on the interaction and docking score, it is evident that pyridine-3-carboxamide had the potential to inhibit the activity of the target receptor ER that could to possibly be used to treat the breast cancer.
The derivatives of pyridine were found to act as have numerous biological activities such as anticancer, antimicrobial, antiviral, antidiabetic and antithrombic agents in the pharamaceuticalindustry 32. As the marine sponge Dysideaherbacea contains pyridine -3-carboxamide which was found to be effective in targeting receptors such as EGFR and ER, further the compound could be isolated and purified and tested on animal models to ascertain as a potential drug to treat breast cancer.
Evaluating the drug-likeness of compound of interest
The LogP of a small molecule is based on the sum of fragment-based contributions and correction factors especially of all organic and organometallic molecules. By measuring the hydrophobicity of the small molecule, the absorption properties, bioavailability, its interaction with the receptor and metabolism of the molecule and its toxicity could be predicted and are used in QSAR studies and rational drug design which measures molecular hydrophobicity.Predicting the polar surface area and molecular volume determines the transport characteristics of specific molecules such as how well the molecule is intestinally absorbed or penetrate blood brain barrier. The volume determines its molecular properties and biological activity that aids in QSAR studies.
The rule of five properties is a set of simple molecular descriptors used by Lipinski, to assess the behavior of small molecule as a drug. The rule states that most drug like molecules have logP =5, molecular weight <=500, number of hydrogen bond acceptors <=10, and number of hydrogen bond donors <=5. Molecules violating more than one of these rules may have problems with bioavailability33.
Conclusion
This study showcases the importance of small molecules identified fromDysideaherbacea and their ligand efficiency on target receptors such as ER and EGFR. The conformation of the docked compounds had shown more binding affinity towards the active site region of the receptor by observing the docking score and ligand efficiency. The findings suggest that the molecules pyridine-3-carboxamide, 1-Chloro-3-(naphthalen-1-yl) propan-2-one and 13-Oxabicyclo tridecane could stands as a drug molecule to treat breast cancer. Further, these compounds could be simulation to understand the structural dynamics and isolated and tested for their anticancer property by performing in-vitro studies.
Acknowledgement
The author Habeeb Shaik Mohideen was earlier publishing articles earlier as S. K. M. Habeeb/Habeeb. S. K. M.
Conflict of Interest
Authors declare no conflict of interest with any institute/individual/organisation.
References
- Zubeda et al., “HER-2/neu Status: A Neglected Marker of Prognostication and Management of Breast Cancer Patients in India,” Asian Pac. J. Cancer Prev., vol. 14, pp. 2231–2235, Apr. 2013.
CrossRef - Shou et al., “Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer.,” J. Natl. Cancer Inst., vol. 96, no. 12, pp. 926–935, Jun. 2004.
CrossRef - D. Yager and N. E. Davidson, “Estrogen carcinogenesis in breast cancer.,” N. Engl. J. Med., vol. 354, no. 3, pp. 270–282, Jan. 2006.
CrossRef - S. Benson, S. D. Babu, S. Radhakrishna, N. Selvamurugan, and B. R. Sankar, “Grade Dependent Expression of Growth Factor Receptors and Signaling Molecules in Breast Cancer,” J. Cancer Ther., vol. 04, no. 07, pp. 21–31, 2013.
CrossRef - Y. Wang and L. Yin, “Estrogen receptor alpha-36 (ER-α36): A new player in human breast cancer,” Mol. Cell. Endocrinol., vol. 418, Apr. 2015.
CrossRef - Kiani, A. Khan, H. Khawar, F. Shuaib, and S. Pervez, “Estrogen receptor α-negative and progesterone receptor-positive breast cancer: Lab error or real entity?,” Pathol. Oncol. Res., vol. 12, no. 4, p. 223, 2006.
CrossRef - Zhang, H. Zhang, X. Wang, J. Wang, X. Zhang, and Q. Zhang, “The eradication of breast cancer and cancer stem cells using octreotide modified paclitaxel active targeting micelles and salinomycin passive targeting micelles.,” Biomaterials, vol. 33, no. 2, pp. 679–691, Jan. 2012.
CrossRef - Shaik, H. Alkreathy, G. Ajabnoor, P. Verma, and B. Banaganapalli, “Molecular Designing, Virtual Screening and Docking Study of Novel Curcumin Analogue as Mutation (S769L And K846R) Selective Inhibitor for EGFR,” Saudi J. Biol. Sci., vol. 26, May 2018.
CrossRef - Yang, A. Roy, and Y. Zhang, “BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions,” Nucleic Acids Res., vol. 41, no. Database issue, pp. D1096–D1103, Jan. 2013.
CrossRef - K. Shiau et al., “The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.,” Cell, vol. 95, no. 7, pp. 927–937, Dec. 1998.
CrossRef - M. Berman et al., “The Protein Data Bank,” Nucleic Acids Res., vol. 28, no. 1, pp. 235–242, Jan. 2000.
CrossRef - Santhi and S. Aishwarya, “Insights from the molecular docking of withanolide derivatives to the target protein PknG from Mycobacterium tuberculosis.,” Bioinformation, vol. 7, no. 1, pp. 1–4, 2011.
CrossRef - Thomas and J.-Å. Gustafsson, “The different roles of ER subtypes in cancer biology and therapy.,” Nat. Rev. Cancer, vol. 11, no. 8, pp. 597–608, Jul. 2011.
CrossRef - F. S Reihani, S. Ramli, A. Alkarkhi, and A. Easa, “Total phenolics, flavonoids and antioxidant activity of banana pulp and peel flours: Influence of variety and stage of ripeness,” Int. Food Res. J., vol. 19, pp. 1041–1046, Jan. 2012.
- B. Dickson and G. M. Stancel, “Estrogen receptor-mediated processes in normal and cancer cells.,” J. Natl. Cancer Inst. Monogr., no. 27, pp. 135–145, 2000.
CrossRef - A. Fuqua, “The role of estrogen receptors in breast cancer metastasis.,” J. Mammary Gland Biol. Neoplasia, vol. 6, no. 4, pp. 407–417, Oct. 2001.
CrossRef - N. Chitrala and S. Yeguvapalli, “Prediction and analysis of ligands against estrogen related receptor alpha.,” Asian Pac. J. Cancer Prev., vol. 14, no. 4, pp. 2371–2375, 2013.
CrossRef - A. Ariazi, J. L. Ariazi, F. Cordera, and V. C. Jordan, “Estrogen receptors as therapeutic targets in breast cancer.,” Curr. Top. Med. Chem., vol. 6, no. 3, pp. 181–202, 2006.
CrossRef - I. Hayashi et al., “The expression and function of estrogen receptor alpha and beta in human breast cancer and its clinical application.,” Endocr. Relat. Cancer, vol. 10, no. 2, pp. 193–202, Jun. 2003.
CrossRef - K. Salih and I. S. Fentiman, “Breast cancer prevention: present and future.,” Cancer Treat. Rev., vol. 27, no. 5, pp. 261–273, Oct. 2001.
CrossRef - S. Herbst et al., “Phase I/II trial evaluating the anti-vascular endothelial growth factor monoclonal antibody bevacizumab in combination with the HER-1/epidermal growth factor receptor tyrosine kinase inhibitor erlotinib for patients with recurrent non-small-cell lung cancer.,” J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol., vol. 23, no. 11, pp. 2544–2555, Apr. 2005.
CrossRef - Al-Kuraya et al., “Prognostic relevance of gene amplifications and coamplifications in breast cancer,” Cancer Res., vol. 64, no. 23, pp. 8534–8540, 2004.
CrossRef - Féger, S. Gil-Falgon, and C. Lamaze, “Cell receptors: definition, mechanisms and regulation of receptor-mediated endocytosis.,” Cell. Mol. Biol. (Noisy-le-grand)., vol. 40, no. 8, pp. 1039–1061, Dec. 1994.
- Fan, Y. Lu, X. Wu, and J. Mendelsohn, “Antibody-induced epidermal growth factor receptor dimerization mediates inhibition of autocrine proliferation of A431 squamous carcinoma cells.,” J. Biol. Chem., vol. 269, no. 44, pp. 27595–27602, Nov. 1994.
CrossRef - L. Arteaga, “The epidermal growth factor receptor: from mutant oncogene in nonhuman cancers to therapeutic target in human neoplasia.,” J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol., vol. 19, no. 18 Suppl, pp. 32S-40S, Sep. 2001.
CrossRef - Stamos, M. X. Sliwkowski, and C. Eigenbrot, “Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor.,” J. Biol. Chem., vol. 277, no. 48, pp. 46265–46272, Nov. 2002.
CrossRef - S. Kim, F. R. Khuri, and R. S. Herbst, “Epidermal growth factor receptor biology (IMC-C225).,” Curr. Opin. Oncol., vol. 13, no. 6, pp. 506–513, Nov. 2001.
CrossRef - F. Allen, P. F. Lenehan, I. A. Eiseman, W. L. Elliott, and D. W. Fry, “Potential benefits of the irreversible pan-erbB inhibitor, CI-1033, in the treatment of breast cancer.,” Semin. Oncol., vol. 29, no. 3 Suppl 11, pp. 11–21, Jun. 2002.
CrossRef - L. Weigel and N. L. Moore, “Cyclins, cyclin dependent kinases, and regulation of steroid receptor action.,” Mol. Cell. Endocrinol., vol. 265–266, pp. 157–161, Feb. 2007.
CrossRef - J. Welboren, F. C. G. J. Sweep, P. N. Span, and H. G. Stunnenberg, “Genomic actions of estrogen receptor alpha: what are the targets and how are they regulated?,” Endocr. Relat. Cancer, vol. 16, no. 4, pp. 1073–1089, Dec. 2009.
CrossRef - Shao and M. Brown, “Advances in estrogen receptor biology: prospects for improvements in targeted breast cancer therapy.,” Breast Cancer Res., vol. 6, no. 1, pp. 39–52, 2004.
CrossRef - Chaubey and S. N. Pandeya, “Pyridine” a versatile nucleuse in pharmaceutical field,” Asian J. Pharm. Clin. Res., vol. 4, no. 4, pp. 5–8, 2011.
- A. Lipinski, F. Lombardo, B. W. Dominy, and P. J. Feeney, “Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings,” Adv. Drug Deliv. Rev., vol. 23, no. 1, pp. 3–25, 1997.
CrossRef