Tasneem Humaid Ahmed Al-Habsi1, Ruwaida Nasser Abdulla AL-Lamki2 and Mohamed Mabruk1
1Department of Allied Health Sciences, College of Medicine and Health Sciences, Sultan Qaboos University, Oman
2Department of Microbiology, College of Medicine and Health Sciences, Sultan Qaboos University, Oman
Corresponding Author E mail: mabruk@squ.edu.om
DOI : https://dx.doi.org/10.13005/bpj/2087
Abstract
Background:Wound infections are considered as a major cause of morbidity and mortality around the world and it is associated with long hospital stays and increased costs. Aims:To determine the prevalence of bacterial pathogens, associated risk factors and the antibiotic susceptibility patterns of wound infections amongpatients attending a tertiary care hospital in Oman. Methods:One hundred and sixty wound swabs were collected using clean, sterile swabs from patients attending Sultan Qaboos UniversityHospital(SQUH), as a tertiary care hospital in Oman. These wound swabswere inoculated into appropriate culture media. A microscopical examination was carried out in order to have a preliminary idea of the causative microorganism untilthe culture reports were available.Bacterial growth was identified by morphological aspects of colonies, followed by different biochemical tests.BD PhoenixTMsystem was used to confirm microbial identification and to determine the antibiotic sensitivities. Results:Out of 160 wound swabs, 93(58.1%) were positive for wound infections.Staphylococcus aureuswas the most prevalent microorganism. Elderly patients above 70 years had more wound infections compared to other age groups. Most of the isolated Gram-positive bacteria were sensitive to vancomycin except Enterococcus gallinarum.Isolated Gram-negative bacteria were 100% resistant to ampicillin except for Proteus mirabilis. Conclusion:Multi-drug resistant (MDR) organisms had quit high prevalence in wound infections among Omani patients, therefore there is a need for adequate intervention to limit the spread and evolution of further resistance.
Keywords
Antibiotic resistance; Oman; Wound infections
Download this article as:| Copy the following to cite this article: Al-Habsi T. H. A, AL-Lamki R. N. A, Mabruk M. Antibiotic Susceptibility Pattern of Bacterial Isolates from Wound Infections Among Patients Attending A Tertiary care Hospital in Oman. Biomed Pharmacol J 2020;13(4) |
| Copy the following to cite this URL: Al-Habsi T. H. A, AL-Lamki R. N. A, Mabruk M. Antibiotic Susceptibility Pattern of Bacterial Isolates from Wound Infections Among Patients Attending A Tertiary care Hospital in Oman. Biomed Pharmacol J 2020;13(4). Available from: https://bit.ly/2KZVAum |
Introduction
A wound is defined as “a breakdown in the protective function of the skin, the loss of continuity of epithelium, with or without loss of underlying connective tissue”1. These wounds range from minor cuts and burns to major surgical wounds and body ulcers1,2.
Pathogen infecting wounds can originate either from the external environment or from the patient’s endogenous flora such as the patient’s skin, mucous membranes, or gastrointestinal tract 3.
Wounds are classified into acute and chronic wounds. An acute wound is usually caused by external damage to the skin which is the case in surgical wounds, burns, bites and minor cuts. While chronic wounds are usually caused by disturbance of the dermal and epidermal tissue by an endogenous mechanism due to a predisposing condition such as diabetic foot ulcers and pressure sores 4.
Wound infections usually occur when the virulence factors of the pathogen overcome the host immune system 5, 6.
The causative agents of wound infections may vary with the geographical location, from hospital to hospital and with different surgical procedures performed7. The most common causative agents of wound infections are Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella pneumoniae and Escherichia coli7.
Many studies around the world have been conducted to identify the bacterial species isolated from wound infections.A study carried out in Italy, showed that the most common bacterial species isolated from different types of wounds were Staphylococcus aureus (37%), followed by Pseudomonas aeruginosa (17%), Proteus mirabilis (10%), Escherichia coli (6%) and Corynebacterium spp. (5%) 8.
A previously, published study carried out in south-west Ethiopia showed the predominance of Staphylococcus aureus (32.4%) followed by Escherichia coli (20%), Proteus species (16%), Coagulase negative Staphylococci (14.5%), Klebsiella pneumoniae (10%) and Pseudomonas aeruginosa (8%) 9.
Another study carried outin Egypt showed that Pseudomonas aeruginosa was the most frequent isolated microorganism from burn infections in cancer patients. While Staphylococcus aureus was the most frequent isolated microorganism from wound infections in cancer patients 10.
A previously published study from Saudi Arabia showed that only 23 patients out of 131 patients admitted to surgical wards and surgical intensive care unit in King Khalid University Hospital had bacteria isolated from their surgical sites11. In addition, emergency operations showed a higher rate of infections in comparison to elective operations. The most common bacterial isolates were Escherichia coli followed by Pseudomonas aeruginosa and Staphylococcus aureus11.
Bacterial wound infections are treated with different types of antibiotics. The selection of the suitable antibiotic depends on a number of factors including the causative agent, the site and severity of the infection 3. The chosen antibiotic should be able to eliminate the microorganism completely and at the same time, cause the least adverse effect by reducing the possibilities of the microorganism to have a tendency to develop antibiotic resistance 3.
Unfortunately, bacteria have developed several mechanisms through which it can resist the action of antibiotics. This includes mutations in genes encoding the target site of the antibiotic, over-expression of efflux pumps that extrude the drug from the cell, and protection of the antibiotic target site by specific proteins 12, 13. Sydnor and Perl(2011)showed that multi drug resistant bacteria had shown a higher mortality rate compared to those caused by susceptible ones14.
In a study conducted in Italy, gram positive bacterial isolates from wound swabs were susceptible to vancomycin and linezolid, while gram negative bacterial isolates showed quite high resistance to most of the tested antibiotics, where amikacin was the most effective against them8.
In a study carried out in Nigeria, all wound isolated microorganisms were resistant to cloxacillin 15. Pseudomonas aeruginosa had a very high resistance to tested antibiotics, with the lowest resistance to ofloxacin, while Klebsiella pneumonia was relatively susceptible to nitrofurantoin15. Multiple-antibiotic resistant strains including Pseudomonas aeruginosa, coliforms and Staphylococcus aureus were resistant to all tested antibiotics15.
This study and for the first time was carried out to firstly identify the causative bacterial pathogens of wound infections among patients attending a tertiary care hospital in Oman and to determine the antibiotic susceptibility patterns of the isolated bacterial pathogens and finally, to investigate the risk factors contributing to wound infections.
Materials and Methods
Specimens
One hundred and sixty wound swabs were collected using clean, sterile swabs from patients attending SQUH followed by adequate labelling of the sample with necessary data (patient’s name, medical record number, episode number, age, gender and date of collection). All patients with suspected wound infections and attended Sultan Qaboos University Hospital in the period from August until November 2018 were included in this present study. Patients on antibiotic therapy 2 weeks before the study was excluded.
To avoid contamination of wound swab samples, all the swabbing was carried out following the hospital guidelines by a well-trained and qualified Medical officer at the Sultan Qaboos University Hospital.
All swabs were sent in Amies transport media immediately after collection to the Microbiology Laboratory at the Hospital for Microbiological analysis. Once samples arrived at the laboratory, they were updated on the Lab Track system and request forms including all patients’ information were printed out.
Ethical approval
The ethical approval for this research was obtained from the Research Ethics Committee College of Medicine and Health Science, Sultan Qaboos University, Muscat, Oman. (MREC#1689).
Microscopical examination of wound swabs.
Wound swabs were smeared on a glass slide, heat fixed on a hot plate and stained with gram stain. A microscopical examination was carried out in order to have a preliminary idea of the causative microorganism until the culture reports were available.
Detection of pathogenic bacteria in wound swabs using routine culture media
Detection of the causative microorganism of wound infection was done by culturing the wound swabs into blood agar, CLED agar, neomycin blood agar and Sabouraud agar. Inoculation of wound swabs into Sabouraud agar was only performed if yeasts were seen duringthe microscopical examination. All plates except neomycin agar were incubated in aerobic conditions for 24 hours at 37°C, while neomycin agar was incubated in 7% CO2 at 37°C for 48 hours after the addition of 5 mg metronidazole (MTZ) disk to the plate. After incubation, the bacterial growth was identified by morphological aspects of the colonies, followed by different biochemical tests such as coagulase, catalase and oxidase tests. The identification of the pathogens was confirmed using automated BD PhoenixTM system.
Identification and antibiotic susceptibility testing of bacterial pathogens in wound swabs using BD Phoenix system
The BD PhoenixTM automated identification and susceptibility system provides accurate, rapid and reliable identification of known and newly emerging antimicrobial resistance (Bd,2019). Bacterial colonies from culture plates were added to ID broth to prepare a 0.5 McFarland suspension with the aid of BD PhoenixTM nephelometer. 25ml of prepared ID broth was added to AST broth in addition to one drop of AST indicator. ID broth and AST broth were inoculated into the panel wells and purity plates were prepared from the inoculum fluid for purity check. Finally, panels were loaded into BD PhoenixTM automated system and reports were printed out when the processing was completed.Manual antibiotic susceptibility testing using disk diffusion method was done for some microorganisms if there was no suitable panel for the identified microorganism. Interpretation of results was done according to the National Committee for Clinical Laboratory Standards (NCCLS) recommendations.
Data analysis
Patients’ information was recorded in the Microsoft Excel program. Statistical analysis was conductedusing both Microsoft Excel program and the Statistical Package for Social Science (SPSS)software in which the mean, median and frequencies for categorized variables were analyzed.
Results
Clinical features
During the data collection period fromAugust untilNovember 2018, a total of 160 patients were recruited. Patients were aged between 0-90 years with a median age of 46 years. Samples were collected from Sultan Qaboos University Hospital (SQUH), which is a referral hospital from all regions of Oman. Out of the 160 patients,87 were males and 73 were females. A wound swab was collected from each patient. The majority of the cases (20%) were 70 years old and above. The sociodemographic characteristics of patients are summarized in Table 1.
Table 1: The sociodemographic characteristics of involved patients.
|
Variables |
Number of cases |
Percentage |
| Gender | ||
| Male | 87 | 54.4% |
| Female | 73 | 45.6% |
| Age | ||
| (median-range) | 46 (0-90) | |
| Age group(years) | ||
| 0-9 | 27 | 16.6% |
| 10-19 | 5 | 3.1% |
| 20-29 | 15 | 9.4% |
| 30-39 | 22 | 13.8% |
| 40-49 | 17 | 10.6% |
| 50-59 | 23 | 14.4% |
| 60-69 | 19 | 11.9% |
| ≥70 | 32 | 20.0% |
Occurrence of pathogens
Out of 160 wound swabs, 93 (58.1%) were positive for wound infections. Out of the 93 positive wound swabs obtained from Omani patients, 47(50.5%) were male and 46 (49.46%) were female. The most abundant isolated pathogenic microorganisms wereStaphylococcus aureus(20.5%), followed by Pseudomonas aeruginosa(13.7%), Klebsiella pneumoniae(9.6%) and Methicillin-Resistant Staphylococcus aureus (MRSA) (5.5%). The prevalence of pathogens detected in wound swabs obtained from Omani patients attending SQUH is summarized in Figure 1.
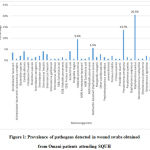 |
Figure 1: Prevalence of pathogens detected in wound swabs obtained from Omani patients attending SQUH |
In about 52 cases (55.9%), the infection was caused by a single microorganism, while in 41 cases (44.1%) the infection was polymicrobial. 75.6 % of the polymicrobial infections were caused by two microorganisms, while 24.4% only were caused by three microorganisms.
The prevalence of bacterial pathogens in male patients
A total of 47 wound swabs obtained from male patients had positive results for bacterial cultures. Staphylococcus aureus was the most common bacterial pathogen isolated from males with 22.2% followed by Klebsiella pneumoniae (11.1%), Pseudomonas aeruginosa (8.3%) and Methicillin Resistant Staphylococcus aureus (MRSA) (5.6%). The type and the percentage of bacterial pathogens detected in wound swabs obtained from male patients are summarized in Figure 2.
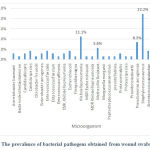 |
Figure 2: The prevalence of bacterial pathogens obtained from wound swabs in male. |
The prevalence of bacterial pathogens in female patients.
A total of 46 wound swabs obtained from female patients had positive results for bacterial cultures. Pseudomonas aeruginosa and Staphylococcus aureus were the most common bacterial pathogens isolated from females with (19.4% each), followed by Klebsiella pneumoniae (8.3%) and Candida albicans (5.6 %). The type and the percentage of microbial pathogens detected in wound swabs obtained from female patients are summarized in Figure 3.
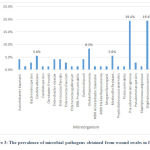 |
Figure 3: The prevalence of microbial pathogens obtained from wound swabs in female |
Association of wound infections with age.
Staphylococcus aureus was the most common bacterial species affecting most of the age groups and it was more associated with patients from (0-9)years old. Whereas Pseudomonas aeruginosa was more associated with patients who were 70 years old and above. Elderly patients above 70 years had more wound infections compared to other age groups. Table 2 shows the distribution of wound infections in association with age groups.
Table 2: The distribution of wound infections in association with age groups
| Microorganism | 0-9 | 10-19 | 20-29 | 30-39 | 40-49 | 50-59 | 60-69 | >=70 |
| Acinetobacter baumanii | 2 | 1 | 2 | |||||
| Acinetobacter baumanniicalcoaceticus complex | 1 | 0 | ||||||
| Bacteroides species | 1 | 2 | ||||||
| Candida albicans | 2 | 1 | 3 | |||||
| Candida species | 1 | 1 | 2 | |||||
| Citrobacter freundii | 1 | 1 | ||||||
| Clostridium species | 1 | |||||||
| Enterobacter aerogenes | 1 | 1 | ||||||
| Enterobacter cloacae | 1 | 2 | 0 | |||||
| Enterococcus faecalis | 1 | 2 | 1 | 2 | ||||
| Enterococcus faecium | 1 | 2 | 1 | |||||
| Enterococcus gallinarum | 1 | |||||||
| Enterococcus raffinosis | 1 | 0 | ||||||
| ESBL Escherichia coli | 1 | 0 | ||||||
| ESBL Klebsiella pneumonia | 1 | 1 | 0 | |||||
| ESBL Proteus mirabilis | 1 | 0 | ||||||
| Escherichia coli | 2 | 1 | 2 | 1 | ||||
| Finegoldia magna | 1 | 0 | ||||||
| Klebsiella pneumonia | 2 | 1 | 3 | 2 | 1 | 3 | 2 | |
| MDR Acinetobacter baumanii | 1 | 0 | ||||||
| MDR Escherichia coli | 1 | 0 | ||||||
| MDR Klebsiella pneumonia | 2 | 0 | ||||||
| Methecillin Resistant Staphylococcus aureus | 2 | 1 | 1 | 2 | 1 | 1 | 0 | |
| Morganella morganii | 1 | 1 | ||||||
| Peptostreptococcusanaerobius | 1 | 1 | 0 | |||||
| prevotellabivia | 1 | 0 | ||||||
| Proteus mirabilis | 1 | 1 | 1 | 0 | ||||
| Proteus species | 1 | 0 | ||||||
| Providencia stuartii | 1 | |||||||
| Pseudomonas aeruginosa | 2 | 1 | 1 | 5 | 2 | 9 | ||
| Pseudomonas species | 2 | 1 | 0 | |||||
| Staphylococcus anginosus | 1 | 0 | ||||||
| Staphylococcus aureus | 10 | 2 | 5 | 3 | 6 | 1 | 3 | |
| Stenotrophomonas maltophilia | 1 | 0 | ||||||
| Streptococcus agalactiae | 1 | 2 | ||||||
| Streptococcus pyogenes | 2 | 0 | ||||||
| Streptoccoccus pneumonia | 1 | 0 | ||||||
| Total | 23 | 3 | 8 | 23 | 8 | 30 | 17 | 35 |
Association of wound infections with the anatomical site.
Twenty-six-point three percent (26.3%) of wound infections were located on the abdomen, followed by breast/chest (18.8%), legs (16.3%), back (12.5%), foot(10.0%),genitalia (8.8%), head and neck (5.0%) and arm (2.5%). Most Staphylococcus aureus caused wound infections were found on the abdomen, whereas most of Pseudomonas aeruginosa caused infections were found on the abdomen and breast/chest area. The distribution of wound infections in association with the anatomical site is summarized in Table 3.
Table 3: The distribution of wound infections in association with the anatomical site.
| Microorganism | Abdomen | Arm | Back | Breast / Chest | Foot | Genitalia | Head and Neck | Leg | unpecified |
| Acinetobacter baumanii | 2 | 1 | 1 | 1 | |||||
| Acinetobacter baumanniicalcoaceticus complex | 1 | ||||||||
| Bacteroides species | 1 | 1 | 1 | ||||||
| Candida albicans | 1 | 1 | 3 | 1 | |||||
| Candida species | 2 | 1 | 1 | ||||||
| Citrobacter freundii | 1 | 1 | |||||||
| Clostridium species | 1 | ||||||||
| Enterobacter aerogenes | 2 | ||||||||
| Enterobacter cloacae | 2 | 1 | |||||||
| Enterococcus faecalis | 2 | 1 | 1 | 1 | 1 | ||||
| Enterococcus faecium | 1 | 1 | 1 | 1 | |||||
| Enterococcus gallinarum | 1 | ||||||||
| Enterococcus raffinosis | 1 | ||||||||
| ESBL Escherichia coli | 1 | ||||||||
| ESBL Klebsiella pneumonia | 2 | ||||||||
| ESBL Proteus mirabilis | 1 | ||||||||
| Escherichia coli | 1 | 1 | 1 | 1 | 2 | ||||
| Finegoldia magna | 1 | ||||||||
| Klebsiella pneumonia | 3 | 2 | 4 | 1 | 2 | 2 | |||
| MDR Acinetobacter baumanii | 1 | ||||||||
| MDR Escherichia coli | 1 | ||||||||
| MDR Klebsiella pneumonia | 2 | ||||||||
| Methecillin Resistant Staphylococcus aureus | 3 | 1 | 1 | 1 | 2 | ||||
| Morganella morganii | 1 | 1 | |||||||
| Peptostreptococcusanaerobius | 1 | 1 | |||||||
| prevotellabivia | 1 | ||||||||
| Proteus mirabilis | 1 | 1 | 1 | ||||||
| Proteus species | 1 | ||||||||
| Providencia stuartii | 1 | ||||||||
| Pseudomonas aeruginosa | 3 | 2 | 3 | 1 | 2 | 2 | 6 | ||
| Pseudomonas species | 1 | 1 | |||||||
| Staphylococcus anginosus | 1 | ||||||||
| Staphylococcus aureus | 7 | 3 | 5 | 3 | 3 | 2 | 5 | 2 | |
| Stenotrophomonas maltophilia | 1 | ||||||||
| Streptococcus agalactiae | 1 | 1 | 1 | ||||||
| Streptococcus pyogenes | 1 | 1 | |||||||
| Streptoccoccus pneumonia | 1 | ||||||||
| total | 34 | 2 | 17 | 20 | 13 | 11 | 8 | 19 | 21 |
Association of wound infections with the wound type.
Wound infections were more associated with surgical wounds (17), followed byabscesses (7) and bedsores (7), traumatic wounds (4), burns (3), diabetic ulcers (3), ulcers (1) and scars (1).
Antibiotic resistance
Most of the isolated Gram-positive bacteria were sensitive to vancomycin except Enterococcus gallinarum which exhibited complete resistanceto vancomycin. All isolated gram-negative bacteria were 100% resistant to ampicillin except Proteus mirabiliswhich was sensitive to it.
Staphylococcus aureus, MRSA and Enterococcus species were completely sensitive to Teicoplanin. Klebsiella pneumonia was completely sensitive to Amikacin, Gentamycin and Trimethoprim/Sulfamethoxazole
Antibiotic resistance of all isolated microorganisms from wound infections are summarized in Table 4 and Table 5.
Table 4: Antibiotic resistance pattern of Gram-positive bacteria to different antibiotics.
| CL | D | ER | G | L | P | TS | TC | T | V | |
| Staphylococcus aureus | 3.2% | 0% | 9.7% | 0% | 0% | 96% | 6.9% | 20.7% | 0% | 0% |
| Enterococcus faecalis | 100% | 0% | 66.7% | 100% | 0% | 100% | 66%% | 0% | 0% | |
| Enterococcus faecium | 100% | 66.7% | 100% | 0% | 100% | 33.3% | 0% | 0% | ||
| Enterococcus gallinarum | 100% | 0% | 0% | 100% | 0% | 100% | 0% | 100% | ||
| Enterococcus raffinosis | 100% | 0% | ||||||||
| MRSA | 22.2% | 0% | 33.3% | 22.2% | 0% | 100% | 25% | 11.1% | 0% | 0% |
| Streptococcus agalactiae | 0% | 0% | 0% | 0% | 0% | 50% | ||||
| Streptococcus anginosus | 0% | 0% | 0% | |||||||
| Streptococcus pneumoniae | 0% | 0% | 0% | 100% | 0% | 0% | ||||
| Streptococcus pyogenes | 0% | 0% | 0% | 0% | 50% | 0% | 0% | |||
| Providencia stuartii | 100% | 100% | 100% |
CL:Clindamycin, D:Daptomycin, ER:Erythromycin, G:Gentamycin, L:Linezolid, P: Penicillin, TS:Trimethoprime/Sulfamethoxazole, TC:Tetracyclin, T:Teicoplanin, V:Vancomycin
 |
Table 5: Antibiotic resistance pattern of Gram-negative bacteria to different antibiotics |
Discussion
In the present study, 93 (58.1%) of the 160 wound swabs collected from patients who attended a tertiary care hospital in Oman for the period from August until November 2018 were positive for wound infections. This finding was to some degree comparable with a study conducted in Italy, where 217 (69.5%) out of 312 wound swabs were positive for wound in fections8.
In our study, the number of infected wounds associated with females 46 (49.5%) was almost the same as males 47(50.5%). These results were consistent with a study conducted in Nigeria15.
In the present study,Staphylococcus aureus was the most predominant bacterial species isolated from wound infections. This result was consistent with the studies conducted in Italy, South-west Ethiopia and Egypt8–10. However, the results of this study were in consistent with the result of another study carried out in Nigeria15, where Pseudomonas aeruginosa was the most commonly detected pathogen15. This could be due to different economic and environmental factors in the two regions.
Pseudomonas aeruginosa was the second most isolated microorganism in the present study. In many studies 8–11,15P.aeroginosa was found to be either the first or second most isolated microorganism from wound infections. Rossi et al., (2015), reported that S.aureus is usually isolated from the superficial layers of wounds, while P.aeroginosa lay in the deepest region of wounds16.
In the present study, most of the wound infections were monomicrobial (55.9%),while (44.1%) of wound infections were polymicrobial. This result was consistent with a study conducted in Italy, where the monomicrobial infections (72.8%) were more frequent than polymicrobial infections(27.2%)8. Our results were also consistent with a study carried out in south-west Ethiopia, where 91.6% of wound infections were monomicrobial, while only 8.4% were polymicrobial infections 9.
It is well known that S.aureus and P.aeroginosa produce many virulence factors that worsen infections and delay healing16. The co-infection of wound with both S.aureus and P.aeroginosais found to be more virulent than a single infection of each microorganism separately16,17.
In the present study, elderly patients who were 70 years old and above had more wound infections compared toother age groups. A similar finding was seen in a previous study carried out in Nigeria, where patients aged between 21-30 years were more prevalent to wound infections15.
The present study showed that the highest number of wound infections were located on the abdomen (26.3%). This result was inconsistent with the results of a previous carried study carried out in south-west Ethiopia, where about 30% of wound infections were located on legs9.
Our results showed that the highest numbers of infections were detected in surgical wounds.This result was inconsistent with studies carried out in Nigeria and South-west Ethiopia, where non-traumatic and traumatic wounds were the highest types of wounds associated with wound infections, respectively 9,15. This could be due to the contamination of surgical instruments with biofilms and inappropriate disinfection of surgical sites18,19,20.
In the present study, we found that all gram negative bacteria exhibited complete resistance to ampicillin except Proteus mirabilis which exhibited no resistance. Mama et al. (2014) found that the isolated gram-negative bacteria were 100% resistant to ampicillin except for Proteus species, where 9% were sensitive to ampicillin9.
In the present study, Methicillin-Resistant Staphylococcus aureus (MRSA) was the fourth (5.5%) most common pathogen isolated from infected wound. This finding is inconsistent with a previously published work where a higher rate of MRSA was detected 21.
In conclusion, our data suggested that the major pathogen that was associated with wound infections among patients attending Sultan Qaboos University Hospital was Staphylococcus aureus. The majority of wound infection cases were diagnosed in elderly patients whose ages were 70 years and above. Polymicrobial infections were recognized in 41 cases (44.1%) of the wound infection cases. For antimicrobial resistance, all isolatedgram-positive bacteria were sensitive to vancomycin except Enterococcus gallinarum which interesting lyexhibited complete resistant to vancomycin. All isolated gram-negative bacteria were 100% resistant to ampicillin except Proteus mirabilis.In the present study, multi-drug resistant (MDR) organisms had quiet high prevalence in wound infections among patients attending a tertiary care hospital in Oman;therefore, there is a need for effective intervention to limit the spread and evolution of further antibiotic resistant bacterial pathogens among this unique a group of patients.
Acknowledgment
We would like to thank the head and all the clinical and technical staff at the Microbiology Department, Sultan Qaboos University Hospital for allowing us to use all of their departmental and laboratory facilities and for all of their support and encouragement. Also, the authors would like to thank SQU ethical committee for the approval of this project and for allowing us to obtain patients data. The authors would like to thank the College of Medicine and Health Sciences, Sultan Qaboos University for funding this project.
References
- Green JW, Wenzel RP. Postoperative wound infection: a controlled study of the increased duration of hospital stay and direct cost of hospitalization. Ann Surg;185:264–8 (1977).
CrossRef - Heggers JP . Assessing and controlling wound infection. Clin Plast Surg;30(1):25-3 (2003).
- Hyonsurk Kim. Wound Infection. Arch Plast Surg;46(5): 484–48(2019).
CrossRef - Swindon, Wiltshire, Bath and north east Somerset Wound Group. Identification, diagnosis and treatment of wound infection. Nurs Stand; 16-22;26(11):44-8 (2011 ).
CrossRef - Haesler E , Swanson T , Ousey K , Carville K. Clinical indicators of wound infection and biofilm: reaching international consensus. J wound care; 2;28(Sup3b):4-12 (2019).
CrossRef - Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev;14:244–69 (2001).
CrossRef - Negi V, Pal S, Juyal D, Sharma MK, Sharma N. Bacteriological profile of surgical site infections and their antibiogram: A study from resource constrained rural setting of Uttarakhand state, India J Clin Diagnostic Res;9(10):DC17-20.(2015).
CrossRef - Bessa LJ, Fazii P, Di Giulio M, Cellini L. Bacterial isolates from infected wounds and their antibiotic susceptibility pattern: Some remarks about wound infection. Int Wound J;12:47–52 (2015).
CrossRef - Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob ;13:14 (2014).
CrossRef - Alharbi SA, Zayed ME. Antibacterial susceptibility of bacteria isolated from burns and wounds of cancer patients. J Saudi Chem Soc;18:3–11(2014).
CrossRef - Khairy GA, Kambal AM, Al-Dohayan AA, Al-Shehri MY, Zubaidi AM, Al-Naami MY, et al. Surgical Site Infection in a Teaching Hospital: A Prospective Study. J Taibah Univ Med;6:114–20 (2011)
CrossRef - Friedman ND, Temkin E, Carmeli Y. The negative impact of antibiotic resistance. Clin Microbiol Infect;22:416–22 (2016).
CrossRef - Munita JM, Arias CA. Mechanisms of Antibiotic Resistance. Microbiol Spectr NIH Public Access;4(2):10 (2016).
CrossRef - Sydnor ERM, Perl TM. Hospital epidemiology and infection control in acute-care settings. Clin Microbiol Rev;24:141–73 (2011).
CrossRef - Pondei K, Fente BG, Oladapo O. Current microbial isolates from wound swabs, their culture and sensitivity pattern at the niger delta university teaching hospital, okolobiri, Nigeria. Trop Med Health; 41:49–53 (2013).
CrossRef - Rossi A, Gallelli L, Settimio UF, Amato B, Grande R, Caroleo B, et al. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus . Expert Rev Anti Infect Ther;13:605–13 (2015).
CrossRef - Dissemond J. Methicillin resistant Staphylococcus aureus (MRSA): Diagnostic, clinical relevance and therapy . J der Dtsch Dermatologischen Gesellschaft;7:544–53 (2009).
CrossRef - Owens CD, Stoessel K. Surgical site infections: epidemiology, microbiology and prevention. J Hosp Infect;70:3–10 (2008).
CrossRef - O’Hara LM, Thom KA, Preas MA. Update to the Centers for Disease Control and Prevention and the Healthcare Infection Control Practices Advisory Committee Guideline for the Prevention of Surgical Site Infection: A summary, review, and strategies for implementation. Am J Infect Control;46:602–9 (2018)
CrossRef - Allegranzi B, Bischoff P, de Jonge S, Kubilay NZ, Zayed B, Gomes SM. New WHO recommendations on preoperative measures for surgical site infection prevention: an evidence-based global perspective Lancet Infect. Dis;16(12):276-287 (2016).
CrossRef - Upreti N, Rayamajhee B , Sherchan PS. Prevalence of methicillin resistant Staphylococcus aureus, multidrug resistant and extended spectrum β-lactamase producing gram negative bacilli causing wound infections at a tertiary care hospital of Nepal. Antimicrob Resist Infect Control;7:121(2018).
CrossRef








