Essam Hassan Mohamed1,2* , Salama Mostafa Abdel-Hafez1,3, Mohamed Mohamed Soliman4,5, Saad H. Alotaibi6, Adel Alkhedaide4 and Mostafa Abdulatif Mostafa1,7
, Salama Mostafa Abdel-Hafez1,3, Mohamed Mohamed Soliman4,5, Saad H. Alotaibi6, Adel Alkhedaide4 and Mostafa Abdulatif Mostafa1,7
1Department of Medical Microbiology, Faculty of Applied Medical Sciences, Taif University, Turrabah, 21995, Saudi Arabia.
2Department of Microbiology, Faculty of Veterinary Medicine, Zagazig University, Zagazig 44519, Egypt.
3Animal Reproduction Research Institute, Immunobiology and Immunopharmacology Unit, Giza, Egypt.
4Clinical Laboratory Sciences Department, Turabah University College, Taif University, Turabah, 29541, Saudi Arabia.
5Biochemistry Department, Faculty of Veterinary Medicine, Benha University, Benha 13736, Egypt.
6Chemistry Department, Turabah University College, Turabah 29541, Taif University, Saudi Arabia.
7Zoology and Nematology Department, Faculty of Agriculture, Al-Azhar University, Cairo, Egypt
Corresponding Author E-mail : esam2005micro@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1978
Abstract
The current study was intended to characterize and specify local MRSA isolates based on its phenotypic and genotypic features. Moreover, the anti-biofilm impact of garlic and lactobacillus biosurfactants against S. aureus was examined. A total of 130 samples and isolates were taken from four sheep farms. Ninety samples were nasal, 10 were lung tissues and the remaining 30 were raw sheep meat from local grocery stores. All isolates were examined for the presence of S. aureus by routine and known microbiological tests. Oxacillin and cefoxitin disc diffusion test were done and findings were confirmed by PCR technology for S. aureus specific genes. Lastly, garlic water extract (gWE) and Lactobacillus biosurfactants were examined for their anti-biofilm activity utilizing a microtitre plate test. Fifteen isolates out of 75 S. aureus were specified MRSA. Concerning the PCR results, the 16S rRNA, the nuc and the mecA genes were detected in 75, 15 and 15 isolates, respectively. Moreover, nuc gene was present in all MRSA isolates (100%). The biofilms formation by S. aureus MRSA was repressed significantly by garlic water extract (gWE) and was dose dependently effect. Same repression was reported by Lactobacilli biosurfactants in dose dependent manner. The prevalence of S. aureus MRSA was confirmed and S. aureus biofilm activity was repressed by gWE and Lactobacilli biosurfactants.
Keywords
Anti-biofilm Activity; Adhesion Molecules; Congo Red; Garlic Extract; PCR; Staph MRSA
Download this article as:| Copy the following to cite this article: Mohamed E. H, Hafez S. M. A, Soliman M, Alotaibi S. H, Alkhedaide A, Mostafa M. A. Characterization and Identification of Methicillin-resistant Staphylococcus aureus (MRSA) Producing Biofilm: Impacts of Garlic Extract and Lactobacillus Biosurfactants. Biomed Pharmacol J 2020;13(3). |
| Copy the following to cite this URL: Mohamed E. H, Hafez S. M. A, Soliman M, Alotaibi S. H, Alkhedaide A, Mostafa M. A. Characterization and Identification of Methicillin-resistant Staphylococcus aureus (MRSA) Producing Biofilm: Impacts of Garlic Extract and Lactobacillus Biosurfactants. Biomed Pharmacol J 2020;13(3). Available from: https://bit.ly/30sgN43 |
Introduction
Staphylococcus aureus (S. aureus) is a typical pathogen, causing a great variety of infections on skin.1 Treatment of S. aureus infections with antibiotics is frequently less powerful to insufficient because of the development of antibiotic-resistance strains, Methicillin-resistant S. aureus (MRSA).2 The virulence of S. aureus pathogen relies upon the generation of several factors. There were 24 anchored cell wall proteins that expressed by S. aureus. These proteins promote S. aureus to cling to extracellular materials, influencing on the invasion of non-phagocytic cells and interference with innate immunity.3
MRSA was isolated and known by followings, hospital related MRSA (HA-MRSA), community-related MRSA (CA-MRSA) and livestock-associated MRSA (LA-MRSA) .4 Nowadays, MRSA strains were isolated from different foods sources, such as poultry, pork, beef, milk and vegetables, concluded that foods may act as source of contamination.5 S. aureus biofilm is more significant virulence factor than any other strains. The development of biofilms on medical devices is a major issue in hospitals, as they can turn into a wellspring of disease.6 S. aureus hinder the first line of defense of in the body known as immune system by building a wide variety of active peptides.7
Garlic (Allium sativum L.) contains active antibacterial substance called allicin, diallyl disulfide and alipin, which is powerful against numerous pathogenic microbes.8,9 S-allyl-L-cysteine sulfoxide (alliin) is a garlic organo-sulfur ingredient with no odor. The allinase enzyme, which is the cysteine sulfoxidelyase, turnsalliin into allicin when garlic is cut. The strong smell, antioxidant and antibacterial functions of garlic are attributed to allicin.10 The main biological, biochemical and antioxidant active substance of fresh garlic is named allicin. Probiotic bacteria, for example, Lactobacillus fermentum and Lactobacillus plantarum, found to repress S. aureus biofilm formation, however the mechanism is not well known. S. aureus genes icaA and icaD expression is engaged with the synthesis of the biofilm matrix.11
The Current study was planned to isolate, characterize and identify local MRSA isolates with regards to genotypic and phenotypic features. Moreover, the activity of garlic water extract (gWE) and lactobacillus biosurfactants against biofilm formation by MRSA isolates were examined.
Materials and Methods
Specimen collection and Identification
A total of 130 samples were isolated and collected from four sheep farms and local grocery stores in Turabah governorate, Taif, Saudi Arabia. The collected samples were divided as following, 90 out of 130 samples were nasal swabs, 10 were lung tissues from recently dead sheep suffering from respiratory symptoms and 30 were raw sheep meat collected from local grocery stores. Collection of samples were ranged from October 2017 till April 2018. All samples obtained from live and dead sheep were obtained by veterinarians the guide lines of Taif University, Saudi Arabia. The collected nasal swabs and lung tissue samples were sub-cultured on tryptic soy broth (Difco) and then incubated for 24 hrs at 37 °C and were maintained on trypticase soy agar, (Difco). The meat samples (raw sheep) were taken and kept at 4 °C prior to investigation. 25 grams of meat samples were homogenized with buffered peptone water (225 ml) and 1:10 serial dilution was done. 0.1 ml of diluted samples were streaked on Baired-Parker (BP) agar (Oxiod) supplemented with egg yolk tellurite emulsion (Oxiod). The plates were then incubated for 24 hrs at 37 °C 12. Isolates were primarily identified by Gram stain, Catalase, Culturing on mannitol salt agar, coagulase test 13 and molecular PCR technology.
After the confirmation of S. aureus from previous routine microbiological methods, isolates were identified for MRSA by cefoxitin and oxacillin disc diffusion test according to NCCLS guidelines, 2002 14. Next, All resistant isolates to oxacillin (1 μg) and cefoxitin (30 μg) discs were confirmed by PCR technology for MRSA.
DNA Extraction of MRSA and Triplex PCR Identification
A triplex PCR technology was used to identify and discriminate the S. aureus from other Staph species and MRSA isolates genotypically.15 Then 500 µl of the broth was centrifuged for 5 minutes at 2000 rpm. For the pellet, 500 μl of DEPC-treated water (DNase-RNase free) was added and flushed in a vortex. After boiling in water bath at 100 ºC for 10 minutes. The rubes were centrifuged at 10000 rpm for 5 min, the supernatant contain bacterial DNA was taken and used as a template for PCR.
PCR (triplex) was used to identify S. aureus from other Staph species and to characterize MRSA genotypically. To detect Staphylococcus spp., 16S rRNA gene was used. To detect S. aureus specific gene, nuc gene was used. The mecA expression as a methicillin resistance specific gene was used. The sequence of sense and antisense primers for each gene was used (Table 1). Exactly 5 ml of the rapid extracted DNA were taken as a template in a 25 μl PCR mixture. The PCR reaction was consisted of 5µL of cDNA, 2 µL of 10 pM of each primer, and 12.5 µL master mix for PCR (Promega Corp, Madison, WI, USA). The volume was adjusted to 25 µL with sterile deionized water. PCR reaction was performed in thermal cycler machine (Bio-Rad T100TM). The primers were synthesized from Macrogen Co, Seoul Korea. The DNA amplification was performed as follows: 94ºC for 4 minutes of initial denaturation; 30 cycles of 94ºC for 50 seconds, 58ºC for 50 seconds and 72ºC for 90 seconds; and a final extension at 72 ºC for 7 minutes. Amplified genes were loaded in 1.5% agarose gel (stained with ethidium bromide). The 756-bp (16S rRNA), 310-bp (mecA) and 279-bp (nuc) amplified DNA fragments were separated by electrophoresis in agarose gel and visualized under UV-light using gel documentation system.
Table 1: Triplex PCR for identified staph genes
| Gene name | Direction (5’-3’) | Gene sequence |
| 16 S rRNA (756-bp) | 16S rRNA-F | AACTCTGTTATTAGGGAAGAACA |
| 16S rRNA-R | CCACCTTCCTCCGGTTTGTCACC | |
| nuc (279-bp) | nuc-F | GCGATTGATGGTGATACGGTT |
| nuc-R | AGCCAAGCCTTGACGAACTAAAGC | |
| mecA (310 bp) | mecA-F | GTAGAAATGACGAACGTCCGATA |
| mecA-R | CCAATTCCACATTGTTTCGGTCTAA |
Congo Red Agar Method for Determination of Biofilm Production (CRA)
To detect biofilm production from MRSA isolates, a specific medium. The medium was composed of BHI; 37 gms/L, sucrose; 50 gms/L, agar agar; 10 gms/L, and congo red stain; 0.8 gms/L was used. Congo red medium was prepared and autoclaved. Then the MRSA isolates were inoculated in plates with the medium and incubated for 24 and 48 hrs at 37°C.16 Black (intense black) were considered strong positive. While dark colonies (black colonies) considered intermediate biofilm producers. Pink colonies mean weak biofilm producers, while the non-biofilm producers strains remain red with smooth appearance. The experiment was carried out three times.17
Microtitre Plate Test (MTP) for Quantification of Slime Production of MRSA
The quantification of biofilm formation of isolated MRSA and typical positive on CRA was done as described before,18 with little modifications. Isolates of MRSA were subcultured on TSB supplemented with 0.25% glucose overnight at 37°C with shaking. About 100 µl were added to microtitre plate (96-well) contains 100 µl of fresh TSB supplemented with 0.25% glucose. The plate was incubated for 24 hrs at 37°C. Then, plates carefully washed by PB (PH 7.4) three times to remove none adherent cells. Then plates were dried in the air and maintained in inverted position before staining. Crystal violet (0.4%) was used to stain the adherent cells with a volume of 100 µl per well for 120 seconds and then left to be air-dried and washed by sterile distilled water three times. Finally, Optical density (OD) was measured using spectrophotometer (Smart Spectm Plus- Bio- Rad-USA) at 570 nm. The slime produced from isolates were classified into 4 categories based on obtained OD, strong slime formers (OD570 ≥3.0), moderately slime formers (OD570 ≥1.5 – 2.0), weak slime formers (OD570 ≥0.5-1.0) and none slime formers were (OD570 ≤ 0.5).
Preparation of Garlic Water Extract (gWE)
Garlic was purchased from local markets in Turabah, Saudi Arabia. Buds were cleaned thoroughly, then rinsed with tap water and left for drying for 24 hrs at room temperature. Fifty grams of garlic was crushed in a sterile grinder to obtain fine powder. Then 100 ml of sterile pure water were added to the powder and was left for overnight, filtration was done using Whatman paper No1. Finally, the extract was concentrated using shaking incubator (Model: TH2-300- SN 170824647- China) at 37°C for 6 days. The obtained concentrated garlic extract was kept in deep freezer till use.19
Isolation of Lactobacillus biosurfactants (LAB) from Yoghurt
LAB was isolated from yoghurt purchased from local markets in Turabah, KSA. Briefly; one gram of yoghurt and 9 mL sterile saline water were mixed. Then serially diluted and immediately plated on LB agar (Difco, Bacton, USA), which is selective for Lactobacillus spp. The plates were kept and incubated anaerobically, at 37 °C for 48 hrs. Catalase-negative, gram-positive and rod-shaped bacilli were considered LAB positive, further confirmation was made by culturing of previous obtained colonies on MRSA agar.20
Anti-Biofilm Activity
The anti-biofilm activity was explained before 21 with some modifications. All isolates of MRSA were sub-cultured overnight with fresh sterile tryptic soy broth (TSB) supplemented with 0.25% (w/v) glucose. Then, 100 μl of cultures of each isolate was transferred to 96-well microtitre plates. A volume of 100 μl of different concentration of garlic extract (12.5, 25, 50 and 100 mg/ml) and LAB biosurfactants (50 and100 mg/ml) were added to each well except positive and negative control wells. Wells were incubated at 37 °C for 48 hrs, then gently washed twice with sterile PBS (PH 7.4).22 Thereafter, the biofilm were fixed using 200 μL methanol for 10 min, then stained with 0.1% crystal violet for about 10 min and rinsed 3 times with distilled water. The adherent cells were dissolved using 200 μL of 33% acetic acid, OD was measured using spectrophotometer (Smart Spectm Plus- Bio- Rad-USA) at 570nm.20 Each experiment was repeated three times. Positive control contains bacterial isolates without any treatment but negative controls contains only tryptic soy broth with 0.5% (w/v) glucose. The current results were expressed in biomass formation inhibition percentage.23
Statistical Analysis
Data are calculated and expressed as means ± standard error for values of MRSA isolates. All data analyzed using ANOVA (analysis of variance) by Bonferroni test for SPSS software version 11.5 for Windows (SPSS, IBM, Chicago, IL, USA). All values less than P < 0.05 wee considered statistically significant.
Results
Isolated S. aureus from nasal swabs, lung tissues of dead sheep and raw sheep meat were tested using agar disc diffusion method to examine the MRSA distribution (Figure 1). The obtained results revealed that 15 isolates (8, nasal; 4, lung and 3, raw sheep meat) were positive S. aureus MRSA specific species.
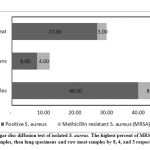 |
Figure 1: Agar disc diffusion test of isolated S. aureus. |
Triplex PCR was used to discriminate S. aureus from other S. species. Triplex PCR targeted the 16S rRNA, the nuc and the mecA genes to be specific genes for Staph, S. aureus and S. aureus specific MRSA, respectively. Detected DNA fragments were with size of 756, 279 and 310 bp, for the 16S rRNA, the mecA and the nuc genes, respectively (Figure 2). From the 130 samples, 75 samples were S. aureus, 15 were positive for both nuc and mecA genes and were proved as MRSA isolates. Strains for S. aureus MRSA were identified by the expression of the mecA gene which confirmed the disc diffusion test results. Moreover, nuc gene was detected in all MRSA isolates (15/15).
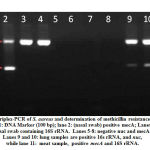 |
Figure 2: Triplex-PCR of S. aureus and determination of methicillin resistance samples. |
CRA test was used for biofilm detection, black colonies with slimy texture revealed positive biofilm MRSA producers, while red colonies indicated none biofilm producing strains (Figure 3A-B). All S. aureus MRSA were cultured on CRA to differentiate between biofilm and none biofilm producers. The obtained results denoted that 12 out of 15 examined MRSA isolates (80%) were considered biofilm producers (black color) meanwhile three isolates (20%) were red color that means negative biofilm producers (Figures 3 and 4). MTP test confirmed the results obtained by CRA. The obtained data was the same as that of CRA as seen in Figure 5.
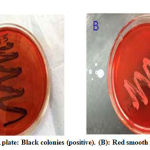 |
Figure 3: A) CRA plate: Black colonies (positive). (B): Red smooth colonies (negative) |
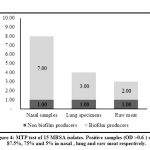 |
Figure 4: MTP test of 15 MRSA isolates. Positive samples (OD >0.6 ) were 87.5%, 75% and 5% in nasal, lung and raw meat respectively. |
S.aureus MRSA biofilms were exposed to garlic extract (gWE). The results of gWE reported that S. aureus MRSA biofilm was inhibited significantly after initial incubation with 25 mg/ml garlic extract (Fig. 5). In parallel, Figure 6 showed the inhibitory effect of Lactobacilli biosurfactants on biofilm generation by MRSA. Compared to control, S. aureus MRSA biofilm was repressed by Lactobacilli biosurfactants at 50 and 100 mg/ml for 18 hrs.
 |
Figure 5: The anti-biofilm activity of garlic water extract (gWE). |
 |
Figure 6: Anti-biofilm activity of Lactobacillus Biosurfactants. |
Discussion
MRSA has gained increasing importance in veterinary medicine in the past 2 decades, as MRSA shows resistance not only β-lactams but also other classes of antimicrobials.24,25 The distribution of MRSA and its several antibiotics resistance are growing worldwide.26 The prevalence of MRSA in large ruminant farms was 34% in milk samples collected from Pakistan.27 The presence of MRSA in samples of meat was previously confirmed,28-30 while it varies greatly by geographical location. Lower results from retail meat (˂ 5%) were reported before, 31,32 while Similar results (10-12 %) were shown in other studies 33,34. Other, confirmed that MRSA prevalence was 31.8% in cattle, 29.8% of sheep, and 11.5% in goats in Jordan.35
Our findings showed that MRSA isolates were detected in 50% of our samples and the difference from other results may be attributed to the origin of samples used as they used milk samples while ours are nasal, lung and raw meat samples. The mecA gene detection by triplex PCR technology is a gold tool to detect MRSA strains compared to conventional disc diffusion test.36 During the past 2 decades, scientists confirmed the increase in prevalence of mecA gene with multiple antibiotics resistance.37
Nearly, all mecA positive isolated S. aureus strains exhibited resistance to oxacillin and cefoxitin. 38 These results go ahead with our reported results, where, all isolated MRSA with mecA gene were all resistant to oxacillin and cefoxitin using disc diffusion test. The nuc gene was found also in all isolated MRSA strains, as it targeted S. aureus specific isolates and was more species specific than 16S rRNA (S. species specific).39 The high percent reported may be attributed to the high use of antibiotics as growth promoters in animals. Same was confirmed in a previous reported study in Jordan (29.8%).35
The biofilms helps the bacterium to resist death and still survive within the host, then cause chronic or persistent infections.40 Biofilm formation plays a vital role in pathogenicity, virulence and antimicrobial resistance to antibiosis.41 The ability of pathogenic bacteria, such as S. aureus, to produce biofilm is the cause for persistent infections.42 Bacteria producing biofilm are resistant to antibiotics, compared with non-biofilm producers.43 The degree of biofilm formation by MRSA depends on the intercellular adhesion molecules production, named intercellular adhesion (ica) loci that include icaA, icaB, icaC, and icaD genes.44 The process of biofilm formation consists of two independent processes: bacteria initially, attached to a solid surface. followed by proliferation and accumulation of bacteria cells, and thus resulted in biofilm formation and maturation.45
There are different methods for phenotypic detection of slime production as CRA which used in our study and considered rapid reliable qualitative test. CRA contains congo red dye that can detect the polysaccharides on the cell wall of both Gram positive and Gram negative bacteria.16 The obtained results showed that (12/15) of MRSA isolates were considered as biofilm producers (black color) (80%). CRA was used in Previous studies and the biofilm producing MRSA ( black color) was confirmed and was with a percentage of 85.1%.46 MTP is another phenotypic test but more convenient and quantitative than CRA depending on measuring of OD using spectrophotometer and can detect S. aureus cell wall polysaccharides adhering to the polystyrene plate directly. The obtained results are coincided with that of CRA. Previous studies showed that (93/94) MRSA isolates were biofilm producers using the microtitre plate test.47
The anti-biofilm impact of water extract garlic (gWE) was investigated. A reduction in biofilm generation was reported at a concentration equivalent to 0.25 mg/ml gWE. The effect of gWE can be attributed to its active components that stifled the biofilm formation.48 In parallel, previous reports confirmed that bio-surfactants separated from Lactobacilli strains has anti-biofilm activity against MRSA. This findings that potentiate our results increase the usage of these natural products as an alternate promising therapies for treatment of S. aureus MRSA strains.49
Conclusions
Current study characterized and determined the prevalence of S. aureus MRSA in Turabah, Saudi Arabia using well established microbiological and molecular techniques. Moreover, confirmed the anti-biofilm activity of garlic and lactobacillus biosurfactants as promising medications for treatment of S. aureus MRSA in animals.
Acknowledgments
We greatly appreciate the contributions of all authors in finishing this study and Deans of Scientific Research Affairs, Taif University, Saudi Arabia for financial support for the project number (Project # 5527-438-1).
Conflict of Interest
The authors declare that no conflict of interests exists for this study
Author Contributions
All authors contributed equally to finish this study. EHM, SAM, MMS were responsible for conception and designed of the study. EHM, MAM and SAM Undertook the isolation and characterization of S. aureus, SHO was responsible for extraction of garlic, AA and MMS undertook the PCR assay, EHM and MAM were responsible for testing of anti-biofilm of garlic and Lactobacillus biosurfactant. Finally, EHM and MMS were responsible for interpretation and analysis of the date obtained. All authors have read and approved the final version of the paper.
References
- David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clinical microbiology reviews.23(3):616-687(2010).
CrossRef - Bagnoli F, Bertholet S, Grandi G. Inferring reasons for the failure of Staphylococcus aureus vaccines in clinical trials. Frontiers in cellular and infection microbiology.2:16(2012).
CrossRef - Kang M, Ko Y-P, Liang X, et al. Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of Gram-positive bacteria inhibit complement activation via the classical pathway. Journal of biological chemistry.288(28):20520-20531(2013).
CrossRef - Petinaki E, Spiliopoulou I. Methicillin-resistant Staphylococcus aureus among companion and food-chain animals: impact of human contacts. Clin Microbiol Infect.18(7):626-634(2012).
CrossRef - Wu S, Huang J, Wu Q, et al. Staphylococcus aureus Isolated From Retail Meat and Meat Products in China: Incidence, Antibiotic Resistance and Genetic Diversity. Front Microbiol.9:2767(2018).
CrossRef - Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nature Reviews Microbiology.12(1):49(2014).
CrossRef - Thammavongsa V, Kim HK, Missiakas D, Schneewind O. Staphylococcal manipulation of host immune responses. Nature Reviews Microbiology.13(9):529(2015).
CrossRef - Kuda T, Iwai A, Yano T. Effect of red pepper Capsicum annuum var. conoides and garlic Allium sativum on plasma lipid levels and cecal microflora in mice fed beef tallow. Food and chemical toxicology.42(10):1695-1700(2004).
CrossRef - Ankri S, Mirelman D. Antimicrobial properties of allicin from garlic. Microbes and infection.1(2):125-129(1999).
CrossRef - Deresse D. Antibacterial effect of garlic (Allium sativum) on Staphylococcu aureus: An in vitro study. Asian J Med Sci.2(2):62-65(2010).
- Melo TA, dos Santos TF, de Almeida ME, et al. Inhibition of Staphylococcus aureus biofilm by Lactobacillus isolated from fine cocoa. BMC microbiology.16(1):250(2016).
CrossRef - BAM F, Hunt J, Abeyta C, Tran T. FDA Bacteriological Analytical Manual. Chapter.7:23(1998).
CrossRef - Holt JG, Krieg N, Sneath PH, Staley J, Williams S. Bergey’s manual of determinative bacteriology. 9th. Baltimor: William & Wilkins.(1994).
CrossRef - Wayne P. Performance Standards of Antimicrobial Susceptibility: National Committee for Clinical Laboratory Standards (NCCLS). NCCLS Approved Standards p100-159.(2002).
CrossRef - Ciftci A, Findik A, Onuk EE, Savasan S. Detection of methicillin resistance and slime factor production of Staphylococcus aureus in bovine mastitis. Braz J Microbiol.40(2):254-261(2009).
CrossRef - Freeman D, Falkiner F, Keane C. New method for detecting slime production by coagulase negative staphylococci. Journal of clinical pathology.42(8):872-874(1989).
CrossRef - Kouidhi B, Zmantar T, Hentati H, Bakhrouf A. Cell surface hydrophobicity, biofilm formation, adhesives properties and molecular detection of adhesins genes in Staphylococcus aureus associated to dental caries. Microbial pathogenesis.49(1-2):14-22(2010).
CrossRef - Gowrishankar S, Duncun Mosioma N, Karutha Pandian S. Coral-associated bacteria as a promising antibiofilm agent against methicillin-resistant and-susceptible Staphylococcus aureus biofilms. Evidence-Based Complementary and Alternative Medicine.2012(2012).
CrossRef - Khan L, Paulino EGM, Lim D, Nadela F, Yadav R, Birring OJSJJoOR. Anti-microbial efficacy of Allium sativum against Streptococcus mutans biofilm formation on orthodontic mini-implants. J Orthod Res.2(3):129(2014).
CrossRef - Tamang JP, Tamang B, Schillinger U, Franz CM, Gores M, Holzapfel WH. Identification of predominant lactic acid bacteria isolated from traditionally fermented vegetable products of the Eastern Himalayas. International journal of food microbiology.105(3):347-356(2005).
CrossRef - Yang B, Lei Z, Zhao Y, et al. Combination Susceptibility Testing of Common Antimicrobials in Vitro and the Effects of Sub-MIC of Antimicrobials on Staphylococcus aureus Biofilm Formation. Front Microbiol.8:2125(2017).
CrossRef - Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods.40(2):175-179(2000).
CrossRef - Cui X, Shi Y, Gu S, Yan X, Chen H, Ge J. Antibacterial and antibiofilm activity of lactic acid bacteria isolated from traditional artisanal milk cheese from Northeast China against enteropathogenic bacteria. Probiotics and antimicrobial proteins.10(4):601-610(2018).
CrossRef - Lee JH. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol.69(11):6489-6494(2003).
CrossRef - Baptiste KE, Williams K, Willams NJ, et al. Methicillin-resistant staphylococci in companion animals. Emerging infectious diseases.11(12):1942(2005).
CrossRef - Vanderhaeghen W, Hermans K, Haesebrouck F, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiology & Infection.138(5):606-625(2010).
CrossRef - Aqib AI, Ijaz M, Anjum AA, et al. Antibiotic susceptibilities and prevalence of Methicillin resistant Staphylococcus aureus (MRSA) isolated from bovine milk in Pakistan. Acta tropica.176:168-172(2017).
CrossRef - Ammar A, Attia A, El-Hamid MA, El-Shorbagy I, El-Kader SA. Genetic basis of resistance waves among methicillin resistant Staphylococcus aureus isolates recovered from milk and meat products in Egypt. Cellular and Molecular Biology.62(10):7-15(2016).
- Darwish WS, Atia AS, Reda LM, Elhelaly AE, Thompson LA, Saad Eldin WF. Chicken giblets and wastewater samples as possible sources of methicillin‐resistant Staphylococcus aureus: Prevalence, enterotoxin production, and antibiotic susceptibility. Journal of food safety.38(4):e12478(2018).
CrossRef - Sergelidis D, Papadopoulos T, Komodromos D, et al. Isolation of methicillin‐resistant Staphylococcus aureus from small ruminants and their meat at slaughter and retail level in Greece. Letters in applied microbiology.61(5):498-503(2015).
CrossRef - Chairat S, Gharsa H, Lozano C, et al. Characterization of Staphylococcus aureus from raw meat samples in Tunisia: detection of clonal lineage ST398 from the African continent. Foodborne pathogens and disease.12(8):686-692(2015).
CrossRef - Ge B, Mukherjee S, Hsu C-H, et al. MRSA and multidrug-resistant Staphylococcus aureus in US retail meats, 2010–2011. Food microbiology.62:289-297(2017).
CrossRef - De Boer E, Zwartkruis-Nahuis J, Wit B, et al. Prevalence of methicillin-resistant Staphylococcus aureus in meat. International journal of food microbiology.134(1-2):52-56(2009).
CrossRef - Boost MV, Wong A, Ho J, O’Donoghue M. Isolation of methicillin-resistant Staphylococcus aureus (MRSA) from retail meats in Hong Kong. Foodborne pathogens and disease.10(8):705-710(2013).
CrossRef - Obaidat MM, Salman AEB, Roess AA. High prevalence and antimicrobial resistance of mecA Staphylococcus aureus in dairy cattle, sheep, and goat bulk tank milk in Jordan. Tropical animal health and production.50(2):405-412(2018).
CrossRef - Sakoulas G, Gold HS, Venkataraman L, DeGirolami PC, Eliopoulos GM, Qian Q. Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. Journal of clinical microbiology.39(11):3946-3951(2001).
CrossRef - Kreausukon K, Fetsch A, Kraushaar B, et al. Prevalence, antimicrobial resistance, and molecular characterization of methicillin-resistant Staphylococcus aureus from bulk tank milk of dairy herds. Journal of dairy science.95(8):4382-4388(2012).
CrossRef - Al-Ashmawy MA, Sallam KI, Abd-Elghany SM, Elhadidy M, Tamura T. Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from milk and dairy products. Foodborne Pathogens and Disease.13(3):156-162(2016).
CrossRef - Unal S, Werner K, DeGirolami P, Barsanti F, Eliopoulos G. Comparison of tests for detection of methicillin-resistant Staphylococcus aureus in a clinical microbiology laboratory. Antimicrob Agents Chemother.38(2):345-347(1994).
CrossRef - Bernardi A, Pizzolitto E, Pizzolitto A. Detection of slime production by coagulase-negative staphylococci isolated from central venous catheter. Rev Cien Farm Apl.28:57-66(2007).
- AlChalabi R, Mohammed R, Adnan R, Al-Bayati S. The Antibacterial Effect of Ethanolic Extract of Allium Sativum on Biofilm Forming Staphylococcus Aureus Which Cause folliculitis. Int J Adv Res.5(5):1175-1182(2017).
CrossRef - Vasudevan P, Nair MKM, Annamalai T, Venkitanarayanan KS. Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Veterinary microbiology.92(1-2):179-185(2003).
CrossRef - Parsek MR, Fuqua C. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. Journal of bacteriology.186(14):4427-4440(2004).
CrossRef - Duran N, Dogramaci Y, Ozer B, Demir C, Kalaci A. Detection of adhesin genes and slime production among Staphylococci in orthopaedic surgical wounds. African Journal of Microbiology Research.4(9):708-715(2010).
- Cerca N, Oliveira R, Azeredo J. Susceptibility of Staphylococcus epidermidis planktonic cells and biofilms to the lytic action of staphylococcus bacteriophage K. Letters in Applied Microbiology.45(3):313-317(2007).
CrossRef - Melo PdC, Ferreira LM, Nader Filho A, Zafalon LF, Vicente HIG, Souza Vd. Comparison of methods for the detection of biofilm formation by Staphylococcus aureus isolated from bovine subclinical mastitis. Brazilian Journal of Microbiology.44(1):119-124(2013).
CrossRef - Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. Journal of microbiological methods.40(2):175-179(2000).
CrossRef - Wu X, Santos RR, Fink‐Gremmels J. Analyzing the antibacterial effects of food ingredients: model experiments with allicin and garlic extracts on biofilm formation and viability of Staphylococcus epidermidis. Food science & nutrition.3(2):158-168(2015).
CrossRef - Sambanthamoorthy K, Feng X, Patel R, Patel S, Paranavitana C. Antimicrobial and antibiofilm potential of biosurfactants isolated from lactobacilli against multi-drug-resistant pathogens. BMC microbiology.14(1):197(2014).
CrossRef








