Manuscript accepted on :29-06-2020
Published online on: 14-07-2020
Plagiarism Check: Yes
Reviewed by: P. Tadi
Second Review by: Dr Ayush Dogra
T. Ruba1, R. Tamilselvi2*, M. Parisa Beham2, N. Aparna2
1Department of ECE, Fatima Michael College of Engineering and Technology, Tamilnadu-625020, India.
2Department of ECE, Sethu Institute of Technology, Tamilnadu-626115, India.
Corresponding Author E-mail : tamilselvi@sethu.ac.in
DOI : https://dx.doi.org/10.13005/bpj/1991
Abstract
Segmentation of brain tumor is one of the crucial tasks in medical image process. So as to boost the treatment prospects and to extend the survival rate of the patients, early diagnosing of brain tumors imagined to be a crucial role. Magnetic Resonance Imaging (MRI) is a most widely used diagnosis method for tumors. Also current researches are intended to improve the MRI diagnosis by adding contrast agents as contrast enhanced MRI provides accurate details about the tumors. Computed Tomography (CT) images also provide the internal structure of the organs. The manual segmentation of tumor depends on the involvement of radiotherapist and their expertise. It may cause some errors due to the massive volume of MRI (Magnetic Resonance Imaging) data. It is very difficult and time overwhelming task. This created the environment for automatic brain tumor segmentation. Currently, machine learning techniques play an essential role in medical imaging analysis. Recently, a very versatile machine learning approach called deep learning has emerged as an upsetting technology to reinforce the performance of existing machine learning techniques. In this work, a modified semantic segmentation networks (CNNs) based method has been proposed for both MRI and CT images. Classification also employed in the proposed work. In the proposed architecture brain images are first segmented using semantic segmentation network which contains series of convolution layers and pooling layers. Then the tumor is classified into three different categories such as meningioma, glioma and pituitary tumor using GoogLeNet CNN model. The proposed work attains better results when compared to existing methods.
Keywords
Magnetic Resonance Imaging; Semantic Segmentation Network
Download this article as:| Copy the following to cite this article: Ruba T, Tamilselvi R, Beham M. P, Aparna N. Accurate Classification and Detection of Brain Cancer Cells in MRI and CT Images using Nano Contrast Agents . Biomed Pharmacol J 2020;13(3). |
| Copy the following to cite this URL: Ruba T, Tamilselvi R, Beham M. P, Aparna N. Accurate Classification and Detection of Brain Cancer Cells in MRI and CT Images using Nano Contrast Agents . Biomed Pharmacol J 2020;13(3). Available from: https://bit.ly/38S9IO4 |
Introduction
The brain is the managing center and is accountable for the execution of all activities throughout the human body. Formation of tumor in brain can threaten the human life directly. The early diagnosis of brain tumor will increase the patient’s survival rate. Among the number of imaging modalities, Magnetic resonance (MR) imaging is extensively used by physicians in order to decide the existence of tumors or the specification of the tumors.1 Magnetic resonance imaging (MRI) contrast agents plays a vital role in diagnosing diseases. This increases the demands for new MRI contrast agents, with an improved sensitivity and superior functionalities. A contrast agent (or contrast medium) is a substance used to increase the contrast of structures or fluids within the body in medical imaging. In the proposed work, the contrast enhanced MRI images are used for analysis. There are four different types of MRI modalities such as T1, T1c, T2 and FLAIR. Fig 1 shows four different types of MRI modalities. These different MRI modalities generate different types of tissue contrast images. Thus it provides valuable information of tumor structure and enabling diagnosis and segmentation of tumors along with their sub regions. Some of the primary brain tumors are Gliomas, Meningiomas, Pituitary tumors, Pineal gland tumors, ependymomas. Gliomas are tumors that develop from glial cells.
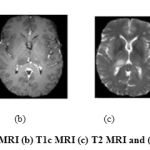 |
Figure 1: (a) T1 MRI (b) T1c MRI (c) T2 MRI and (d) FLAIR |
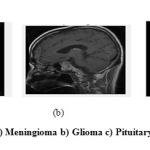 |
Figure 2: a) Meningioma b) Glioma c) Pituitary tumor |
Meningioma often occur in people between the ages of 40 and 70. They are mostly common in women than men. The early diagnosis of brain tumor will increase the patient’s survival rate. In the proposed work three different types of tumors going to be identified such as glioma, meningioma and pituitary tumor. Sample tumor images are shown in Figure 2. The aim of segmentation of brain images is to extract and study anatomical structure and also to identify the size, volume of brain tumor. This helps to predict the increase or decrease the size with treatment. Manual segmentation of MR brain images is time consuming task.2 So that, the automated segmentation methods with high accuracy is mainly focused in research. In current scenario, there are many methods have been developed for automatically segmenting the tumor extents from brain MRI images. Recently, an extremely flexible machine learning approach known as deep learning has emerged as an upsetting technology to enhance the performance of existing machine learning techniques. CNNs (Convolutional Neural Network), are hastily gaining their attractiveness in the computer vision community.
A computed Tomography image shows the internal structure of the bones, organs and tissues. CT scan uses X-rays to provide the structure details. CT images are used for accurate classification of tumor.
Some of the CT images are shown in the Figure 3.
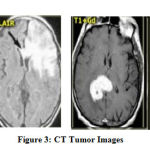 |
Figure 3: CT Tumor Images |
The paper is structured as follows: Section 2 conferred the relevant works in the automated brain tumor segmentation. The attention is mainly on machine learning and deep learning based approaches. The proposed semantic segmentation network for segmentation and GoogLeNet for classification is detailed in section3. Section 4 dealt with the experimental results of the proposed method followed by conclusion in section 5.
Relevant Works
Brain image segmentation is one of the critical tasks for engineers to segment the elements of brain such as white matter, grey matter, tumor etc. In previous decades machine learning plays major role in developing automated brain tumor segmentation. Wei Chen Liu et.al.,3 proposed a novel method for brain tumor segmentation based on features of separated local square. The proposed method basically consists of three steps: super pixel segmentation, feature extraction and segmentation model construction. They use brain images from BRATS 2013 datasets. Also PhamN.H. Tra et.al.,4proposed segmentation method to segment the tumor structure from brain MRI images. For that the images are enhanced using unsharp approach before segmentation using the Otsu method. Similarly Aastha Sehgal et.al.,5 proposed a fully automatic method to detect brain tumors. This method consists of five stages, viz., Image Acquisition, Preprocessing, Segmentation using Fuzzy C Means technique, Tumor Extraction and Evaluation. The improved version of fuzzy c-Means clustering and watershed algorithm has been implemented by Benson.C et.al.6 In that they proposed an effective method for the initial centroid selection based on histogram calculation and then they used atlas-based marker in watershed algorithm for avoiding the over-segmentation problem. Further Boucif Beddad et.al.,7 proposed the notion of FCM which incorporates the spatial information to get a better estimation of the clusters centers. The results obtained are considered as an initialization of the active edge for the Level set. During the application of this method, multiple initializations have been carried out to set the initial parameters.
Though some of the methods discussed above were validated and demonstrated good performance, these fully automated methods for brain tumor detection has not been extremely used in clinical applications. This represents that the detection methods still needs some major developments. Also the machine learning approaches are complex. This growths the development of deep learning based methods for tumor segmentation and classification. Hossam.H et.al.,8 proposed a deep learning (DL) model based on Convolutional Neural Network (CNN) to classify different brain tumor types using publicly available datasets. The former one classifies tumor into (Meningioma, Glioma, and Pituitary Tumor). The other one differentiates between three glioma grades (Grade II, Grade III and Grade IV). Fine tuning of already available CNN can reduce the training time required to develop needed model. So, Guotai et.al.,9 proposed image-specific fine tuning to make a CNN model adaptive to a specific test image, which can be either unsupervised (without additional user interactions) or supervised (with additional scribbles). They also propose a weighted loss function considering network and interaction-based uncertainty for the fine tuning. In the same way, Wang et.al.10 proposed an automatic brain tumor segmentation algorithm based on a 22-layers deep, three-dimensional Convolutional Neural Network (CNN) for the challenging problem of gliomas segmentation. For segmenting tumor, one of the simplest approaches is to train the CNN in Patch-Wise method. First of all an NxN patches are extracted from the given images. Then the model is trained on these extracted patches and produces the labels to each class for identifying the tumor correctly. Most of current well-liked approaches11, 12, 13, 14 using this type of patch wise architectures for segmenting brain tumor. Even more some of the architectures15, 16 provide prediction for every pixel of the entire input image like semantic segmentation.
In the DL based methods discussed in,8, 11-14 the segmentation is fully based on computationally complicated convolution neural network. They design CNN architecture by combining the intermediate results of several connected components and then they classify the images into different categories. They used image patches obtained from axial, coronal, and sagittal views to respectively train three segmentation models, and then combine them to obtain the final segmentation results. The main constraints of these existing methods are expensive computational cost, memory requirements and computational time. In order to provide a solution to the problem mentioned, a modified semantic segmentation networks (CNNs) based method has been proposed.
Proposed Work
As already discussed that the patch wise prediction is complicated and increases the memory and time, in this work a novel hybrid semantic segmentation network for tumor segmentation is proposed. The major contribution of this work is to combine both CNN and semantic segmentation for precise segmentation of tumor region. The complete steps involved in the proposed work for Semantic segmentation and tumor classification using contrast enhanced Brain MRI is given in fig 3. In order to verify the effectiveness of the proposed method, a series of comparative experiments are conducted on the fig share database. To balance the contrast and intensity ranges between patients and acquisition, intensity normalization as preprocessing was performed on each modality. Then the preprocessed brain images are segmented using hybrid semantic segmentation network with series of convolution layers and pooling layers. Then after segmenting the tumor region, the segmented brain images are also classified into one of the three types of tumor such as meningioma, glioma and pituitary tumor using already trained GoogLeNet classifier. Finally, the performance of semantic segmentation and GoogLeNet classification was done using some performance matrices.
Contrast Enhanced MRI Dataset
Figshare dataset is used for evaluating the proposed brain tumor segmentation network.22 This brain tumor dataset containing 3064 T1-weighted contrast-enhanced (T1c MRI) images from 233 patients It includes three kinds of brain tumor such as Meningioma (708 slices), Glioma (1426 slices) and Pituitary tumor (930 slices). The choice of using contrast enhanced MRI is a major part in this research, because in normal MRI there may be limited sensitivity in the probe. This issue can be addressed and rectified by injecting contrast agents through IV (IntraVein) line before taking MRI. Detection of tumor from contrast enhanced MRI will improve sensitivity, specificity and accuracy of the overall system. So that in the proposed work, contrast enhanced MRI and CT images are used. The detailed description of the dataset is shown in table 1. The overall flow diagram is shown in the Figure 4.
Table 1: Detailed description of Figshare dataset
| Tumor Category | Number of Patients | Number of Slices |
| Meningioma | 82 | 708 |
| Glioma | 91 | 1426 |
| Pituitary | 60 | 930 |
| Total | 233 | 3064 |
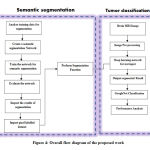 |
Figure 4: Overall flow diagram of the proposed work |
Preprocessing
T1c Magnetic Resonance Imaging is a very useful medical imaging modality for the detection of brain abnormalities and tumor. It may be affected by external noises and also its contrast and intensity are also affected by external environment. So some preprocessing is required to remove unwanted noises and to improve the contrast of the images. In this work, the bias field correction used in23 was adopted to uniform the contrast of the entire images. Also in many applications, color information doesn’t help us to identify important edges or other features. Hence both the MRI and CT images are converted into gray scale images. The input images, its bias field and intensity normalized images are shown in Figure 5.
 |
Figure 5: Sample input image, its bias field and Bias corrected image |
This figure 5 clearly shows that the normalized image has even intensity distribution in every pixel when compared to its original image. Especially in the intensity normalized image, the tumor region is clearly visible.
Hybrid Semantic Segmentation Networking
As described in,19, 25 the main aim of semantic segmentation is to classify the image by labeling each and every of an image. Since each pixels in the images are predicted as some classes, this type of classification is generally referred to as dense prediction. Example for semantic segmentation is shown in figure 6. Machines can augment analysis performed by radiologists, greatly reducing the time required to run diagnostic tests. So in the proposed work, as shown in figure 7, first CNN performs tumor segmentation and the result is going to be overlaid with semantic segmentation to correct the misclassification of pixels. The use of combined semantic segmentation and CNN enables us to perform pixel semantic predictions accurately. Some of the sample images of semantic segmentation are shown in figure 8. Also the layers in CNN for tumor segmentation are shown in figure 9.
 |
Figure 6: Example for semantic segmentation networking |
 |
Figure 7: Hybrid Semantic segmentation network |
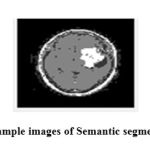 |
Figure 8: Sample images of Semantic segmentation |
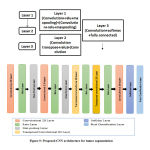 |
Figure 9: Proposed CNN architecture for tumor segmentation |
The proposed CNN model used for tumor segmentation is simple and computationally easier when compared to other architectures. The detailed architecture of proposed CNN is shown in figure 9. The CNN architecture includes 13 layers which starts from the input layer that can hold the images from the previous pre-processing step and then it passes to the convolution layers and their ReLU activation functions which is mainly used for feature extraction. From these features, most probable features were selected using pooling layers. Then down-sampling (convolution, Rectified Linear Unit (ReLU), normalization and pooling layers) was performed. In order to predict the output softmax layer was used and finally a classification layer which produces the predicted class that is tumor and non-tumor regions are segmented from the images.
Finally at the end of these layers the segmented image is obtained which contains tumor and non-tumor regions in the images. After that the output was overlaid with the semantic segmentation. Finally this hybrid segmentation provides accurate segmentation of brain tumor regions. Then this pixel labeled dataset were given as input to GoogLeNet classification for further classification of tumor into three different categories as mentioned earlier.
GoogLeNet Classification
Brain tumor classification is the final step of the proposed approach which is used to identify the type of brain tumor based on the GoogleNet CNN classifier. In the proposed work already trained 22 layer deep GoogLeNet architecture20, 24 is used for classification of tumors. GoogLeNet has a quite different architecture. It uses combinations of inception modules, each including some pooling, convolutions at different scales and concatenation operations. GoogleNet CNN is a feed forward neural networks (FNNs), composed of input and output layers, as well as a single hidden layer. Initializing the weights and biases of the input layer is randomly selected before going to compute the weights of the output layer. The concept of GoogleNet CNN is to classify multiclass problem in more detail. The trained GoogleNet CNN model is utilized to classify the type of brain tumor in an effective manner.
Experimental Results and Discussions
In order to verify the effectiveness of the proposed method, a series of comparative experiments are conducted on the contrast enhanced T1c MRI images of fig share database. In the proposed work, first semantic segmentation is performed followed by CNN with convolution and pooling layers. By inspecting the results of semantic segmentation, the pixel labeled dataset was imported to CNN for further segmentation. Then the images after semantic segmentation was given as input to the CNN with several convolution and pooling layers in which the tumor is segmented separately from brain MRI and CT images. The segmented results for given sample input images of brain tumor are shown in fig 10.
In the proposed method the dataset is divided into two segments. One is for training and other one is for testing sets along with its individual target labels (68% for training and 32% for system test). The network parameters which are set during training are shown in table 2. This table shows different network parameters for segmentation network and classification network. Using these parameters only the network provides accurate results.
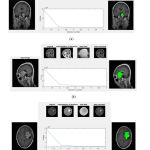 |
Figure 10: Input, bias corrected and Segmentation results |
Table 2: Network parameters for segmentation and classification
| S.No | Factor(s) | Segmentation
Network |
Classification
Network |
| 1 | Optimization method | SGDM | SGDM |
| 2 | Maximum epoch | 50 | 80 |
| 3 | Learning rate | 1e-3 | 1e-2 |
| 4 | Minimum batch size | 4 | 12 |
| 5 | Pooling layer window size | 2 | 3 |
| 6 | Drop factor | 0.1 | 0.1 |
(SGDM – Stochastic Gradient Descent Method)
This figure shows the input images and its corresponding preprocessed images, semantic segmented images and finally tumor segmented images. When we compare the segmented results using proposed method with its ground truth, the proposed segmentation results closely matched with its ground truth images. The performance of the process is measured by calculating the performance metrics of the classifier and segmentation such as Accuracy, Sensitivity and Specificity. These parameters can be calculated using the the formulae as described in.21 The accuracy represents the efficiency of the process. The Sensitivity shows how the algorithm gives correct classification. The Specificity shows how the algorithm rejects the wrong classification results. These parameters can be calculated using equation 1, 2 and 3 which has been taken from. 21
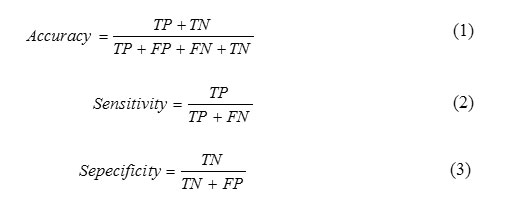
Where, TP-True Positive, TN-True Negative, FP-False Positive and FN-False Negative
The performance of the proposed system is compared with the performance of existing system.6, 17, and 18 The table 3 shows comparison of proposed method with existing method based on classification performance measures.
Table 3: Performance comparison of proposed method
| Methods | Tumor Type | Precision | Sensitivity | Specificity | Accuracy |
| Hossam H. et.al., [6]
|
Meningioma | 0.958 | 0.955 | 0.987 | 97.54% |
| Glioma | 0.972 | 0.944 | 0.951 | 95.81% | |
| Pituitary | 0.952 | 0.934 | 0.97 | 96.89% | |
| Sunanda
Das et.al [17] |
Meningioma | 0.940 | —- | —- | 94.39% |
| Glioma | 0.880 | —- | —- | 94.39% | |
| Pituitary | 0.980 | —- | —- | 94.39% | |
| Hasan
Ucuzal et.al [18] |
Meningioma | 0.945 | 0.877 | 0.986 | 96.29% |
| Glioma | 0.969 | 0.953 | 0.968 | 96.08% | |
| Pituitary | 0.916 | 0.992 | 0.962 | 97.14% | |
|
Proposed Method With MRI Images |
Meningioma | 0.990 | 0.990 | 0.997 | 99.57% |
| Glioma | 0.995 | 0.995 | 0.998 | 99.78% | |
| Pituitary | 0.986 | 0.995 | 0.995 | 99.56% | |
| Proposed Method All
With CT Images |
0.994 | 0.996 | 0.998 | 99.6% | |
In this table, the performance metrics are compared for each tumor types. This table clearly shows that the proposed system provides superior results when compared to existing methods6, 17, and 18 in all aspects of performance. The accuracy of the proposed method is 99.57%, 99.57% and 99.67% for meningioma, glioma and pituitary tumor respectively. Overall, the proposed classification followed by segmentation works very well on the used Figshare dataset. The misclassification of tumor is avoided when compared to already existing tumor segmentation methods. Because in the methods which are already existing for tumor segmentation only uses CNN or any machine learning approaches for tumor segmentation. But in the proposed work pixel based classification named semantic segmentation is combined with CNN for precisely segmenting the tumor. Since each pixels are grouped based on its pixel values, the tumor regions are segregated from the normal healthy tissues. So it becomes easier job for CNN to accurately segment the tumor without any misclassification. Due to that reason only, the proposed work yield almost above 99 % results for each category. The proposed method on CT images also gave better accuracy. Figure 11 shows the plot of classifier performance parameters for each cases of meningioma, glioma and Pituitary tumor for proposed and existing methods.6, 17 and 18 As our method uses modified semantic segmentation networking for segmentation and GoogLeNet for classification, it scores max value in each parameter like precision, sensitivity, specificity and accuracy. This gives max values in all aspects when compared to existing methods. So the proposed work may help the radiologists to take right decision about the treatment of particular tumor. This proposed work can also be extended to taken into account the tumor size and volume in future for the betterment of patient’s treatment.
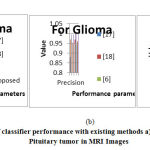 |
Figure 11: Comparison of classifier performance with existing methods |
Conclusion
The proposed method successfully segments the tumor using semantic segmentation network and classifies different tumor categories accurately using GoogLeNet. The quantitative results show that the proposed segmentation and classification yields overall accuracy of 99.57%, 99.78% and 99.56% for meningioma, glioma and pituitary tumors. Due to the use of T1c MRI images and proposed modified semantic segmentation network, the method yield highest possible results in terms of accuracy, sensitivity and specificity. In CT images also the proposed methodology shows better result. All the provided parameters are set properly in MATLAB environment. The performance of semantic segmentation and GoogLeNet classification was done using some performance matrices. This system can even segment non homogeneous tumors providing the non-homogeneity inside the tumor region due to the use of semantic segmentation. The quality of the proposed segmentation was similar to manual segmentation and will speed up segmentation in operative imaging. In future the classifier may be modified to reduce complexity in architecture of GoogLeNet. This proposed work can also be extended to taken into account the tumor size and volume in future for the betterment of patient’s treatment. Also the network can be trained to categorize some other tumor classes such as higher grade glioma (HGG) and lower grade glioma (LGG) with some fine tuning incorporated in the network.
Acknowledgment
“The author/s thankfully acknowledge(s) the financial support provided by The Institution of Engineers (India) for carrying out Research & Development work in this subject”.Grant ID: RDDR2018003
Conflict of Interest
The authors have no Conflicts o interest.
References
- N. Louis et al., “The 2007 who classification of tumors of the central nervous system,” Acta Neuropathologica, vol. 114, no. 2, pp. 97-109, 2007.
CrossRef - Bauer et al., “A survey of MRI-based medical image analysis for brain tumor studies,” Phys. Med. Biol., vol. 58, no. 13, pp. 97-129, 2013.
CrossRef - Wei Chen, Xu Qiao, Boqiang Liu, Xianying Qi, Rui Wang, Xiaoya Wang,” Automatic brain tumor segmentation based on features of separated local square”, Proceeding of Institute of Electrical and Electronics Engineers (IEEE), 2018.
CrossRef - Pham N. H. Tra, Nguyen T. Hai, Tran T Mai,” Image Segmentation for Detection of Benign and Malignant Tumors”, 2016 International Conference on Biomedical Engineering (BME-HUST), 2016.
CrossRef - Aastha Sehgal, Shashwat Goel, Parthasarathi Mangipudi, Anu Mehra, Devyani Tyagi,” Automatic brain tumor segmentation and extraction in MR images”, Conference on Advances in Signal Processing (CASP),2016.
CrossRef - C. Benson, V. Deepa, V. L. Lajish, Kumar Rajamani,” Brain tumor segmentation from MR brain images using improved fuzzy c-means clustering and watershed algorithm”, International Conference on Advances in Computing, Communications and Informatics (ICACCI),2016.
CrossRef - Boucif Beddad, Kaddour Hachemi,” Brain tumor detection by using a modified FCM and Level set algorithms”, 4th International Conference on Control Engineering & Information Technology (CEIT), 18 May 2017.
CrossRef - H. Sultan, Nancy M. Salem, Walid Al-Atabancy,” Multi-Classification of Brain Tumor Images Using Deep Neural Network”, IEEE Access, Volume No: 7, 2019.
CrossRef - Guotai Wang, Wenqi Li, Maria A. Zuluaga, Rosalind Pratt, Premal A. Patel, Michael Aertsen, Tom Doel, Anna L. David, Jan Deprest, Sebastien Ourselin, Tom Vercauteren,” Interactive Medical Image Segmentation Using Deep Learning with Image-Specific Fine Tuning”, IEEE Transactions on Medical Imaging, Volume No: 37, Issue: 7, July 2018.
CrossRef - Wang Mengqiao, Yang Jie, Chen Yilei, Wang Hao,” The Multimodal Brain Tumor Image Segmentation Based on Convolutional Neural Networks”, 2nd IEEE International Conference on Computational Intelligence and Applications (ICCIA), 2017.
CrossRef - Kamnitsas K, “Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation”, Medical Image Analysis, vol. 36, pp.61–78, 2016.
CrossRef - Pereira S, Pinto A, Alves V, Silva CA, “Brain Tumor Segmentation using Convolutional Neural Networks in MRI Images”, IEEE Transaction in Medical Imaging, 2016.
CrossRef - Havaei M, “Brain tumor segmentation with deep neural networks”, Medical Image Analysis, vol.35, pp.18–31, 2016.
CrossRef - Zhang W, “Deep convolutional neural networks for multimodality isointense infant brain image segmentation” Neuroimage, vol.108, pp.214–224, 2015.
- Nie D, Dong N, Li W, Yaozong G, Dinggang S, “Fully convolutional networks for multi- modality isointense infant brain image segmentation”, 13th International Symposium on Biomedical Imaging (ISBI), 2016.
CrossRef - Brosch, “Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation”, IEEE Transaction on Medical Imaging, vol. 35, pp. 1229–1239, 2016.
CrossRef - Sunanda Das, O. F. M. Riaz Rahman Aranya, Nishat Nayla Labiba,“Brain Tumor Classification Using Convolutional Neural Network”,2019 1st International Conference on ICASERT,2019.
- Hasan Ucuzal, ŞeymaYasar, Cemil Çolak, “Classification of brain tumor types by deep learning with convolutional neural network on magnetic resonance images using a developed web-based interface”,2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT),2019.
CrossRef - Shelhamer, J. Long, and T. Darrell, “Fully convolutional networks for semantic segmentation”, IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, no. 4, pp. 640–651, 2017.
CrossRef - Haripriya, R.Porkodi, “Deep Learning Pre-Trained Architecture of Alex Net and Googlenet for DICOM Image Classification”, International Journal of Scientific & Technology Research, Vol. 8, Issue 11, Nov 2019.
- Alireza Baratloo, Mostafa Hosseini, Ahmed Negida, Gehad El Ashal, “Part 1: Simple Definition and Calculation of Accuracy, Sensitivity and Specificity”, Emergency, Vol.3(2), pp. 48-49, 2015.
- Cheng Jun, “brain tumor dataset”, figshare. Dataset, 2017, Available: https://doi.org/10.6084/m9. figshare.1512427.v5.
- J. Tustison, B. B. Avants, P. A. Cook; Y. Zheng, A. Egan, P. A. Yushkevich, and J. C. Gee, “N4ITK: Improved N3 bias correction,” IEEE Transaction on Medical Imaging, vol. 29, no. 6, pp. 1310_1320, Jun. 2010.
CrossRef - Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, and A. Rabinovich, “Going deeper with convolutions,” in The IEEE Conference on (CVPR), June 2015.
CrossRef - Liu, X., Deng, Z. & Yang, Y. Recent progress in semantic image segmentation. Artif Intell Rev,Vol. 52, pp.1089–1106, 2019 Avilable in: https://doi.org/10.1007/s10462-018-9641-3.
CrossRef







