Microbiology Department, College of Medicine , Al Nahtain University, Baghdad, Iraq
Corresponding Author E-mail : dr_maysaa@yahoo.com
DOI : https://dx.doi.org/10.13005/bpj/1970
Abstract
Pyelonephritis-associated pili (Pap) fimbria considered as the main adhesive virulence factor that enable Escherichia coli(E. coli) to colonize in the urinary tract and resist the avoiding by the flow of urine. DNA adenine methyl-transferase gene (Dam) have a role in regulation of papE expression and in bacterial DNA repair system and it could be targeted by antibiotics. Sixty Four isolates of E. coli from urine specimens were obtained from hospitalized and out-patients suffering from signs and symptoms of UTI. These isolates were identified molecularly as uropathic E. coli (UPEC) by detection of papE using PCR. Partial sequencing of pap E was done to study variation among isolates according this gene and its role in susceptibility to antibiotic. Also, Dam was detected using PCR. Detection of papE in E. coli strains revealed that 26/64(42.6%) were considered as UPEC. Analysis of nucleotide sequence changes from partial sequencing tree of pap E shown that there were three clads and UPEC included in clade B displayed the most nucleotide sequence changes. Dam was detected in 11/64 (17.1%) E. coli isolates. The study of multi-drug resistance(MDR) risk in association with the presence of pap E and Dam in UPEC revealed that Dam could be considered as etiological factored to developing MDR. In conclusion, Dam should be taken in consideration as one mechanism of MDR development in UPEC.
Keywords
Escherichia Coli; Multidrug Resistance; Pap E; Dam; Urinary Tract Infection
Download this article as:| Copy the following to cite this article: Dhahi M. A. R . Sequencing Analysis of Pyelonephritis-Associated Pili Gene of Uro-Pathogenic E. Coli Isolated in From Patients-Baghdad, Iraq. Biomed Pharmacol J 2020;13(2). |
| Copy the following to cite this URL: Dhahi M. A. R . Sequencing Analysis of Pyelonephritis-Associated Pili Gene of Uro-Pathogenic E. Coli Isolated in From Patients-Baghdad, Iraq. Biomed Pharmacol J 2020;13(2). Available from: https://bit.ly/2Nzttjn |
Introduction
Uropathogenic Escherichia coli (UPEC) is the most abundant causative bacteria of urinary tract infections (UTIs), worldwide accounting for 80–90% of all infections.1 There are differences in prevalence of virulence factors among UPEC including the adhesins (Type 1, P, S, and F1C fimbriae), pore-forming hemolysin, toxins (RTX and CNF-1),sideophore and aerobactin,which exist in clusters, small virulence cassettes, or large blocks of genes in UPEC strains but, mainly, not observed in the genome of fecal isolates.2,3 Their expression and co-regulation depend on host response and environmental signals.4Iraqi studies from different cities referred to that the frequency of isolation of E. coli from urine of patients with UTI were ranged from 39%-73.6%. 5,6,7,8
The crucial step for starting of UTI is by adhesion of UPEC to mucosal cells that intermediated by P fimbriae and Type-1 fimbriae. Mannose-resistant pyelonephritis-associated pili fimbriae expressed from pap in about 80% of UPEC cause pyelonephritis.9 Genotyping of pap operon referred to its involve fimbriae structural subunits (pap A, C, D,E, F and G) to finish fimbrial growth and attach mature fimbriae to surface of host cell, pap H; and papB and papI ,the divergently encoded regulatory genes, within which the main promoter is located.10
These promoters expressed depending on the DNA methylation of the sequences GATC that located within the intergenic region. DNA methylation is essential epigenetic, post-replicative modification that is catalyzed by a group of enzymes as the DNA methyltransferases (MTases). It is key regulation mechanism of many cellular processes in prokaryotes and eukaryotes. Bacterial DNA methyl-transferases generate N4-methyl-cytosine, C5-methyl-cytosine, or N6-methyl-adenosine in GATC sequences, in an S-adenosyl-methionine-dependent reaction.11,12
DNA adenine methylase (Dam) have important functions in DNA mismatch repair, regulation of transcription, and SOS response activation as part of the cell cycle.13 Methyl-directed mismatch repair is a regulatory process whereby it recognizes the biosynthetic error during the occurrence of the replication fork. The hemi-methylated site differentiates of the DNA template strand and the DNA newly synthesized strand allowing protein MutS to bind to the site where the mismatch occurs and promotes the process of the recruitment of the addition repair proteins to form a ternary complex that initiate repair.13,14 Adherence is essential for UPEC strains, as it avoid removal of bacteria through micturition, and multiple phase-variable adhesins are exists to permit binding to different host tissues. The pap epigenetic switch activates through establishment of differential methylation patterns that regulate the expression phase (on or off) of the pap operon.15,16 Novel antibiotics drug that targets Dam can be intriguing as the enzymatic activity is a lack in human. Inhibiting Dam by DNA methyl-transferase inhibitors (DNMTi) can be detrimental to the bacterium. The inhibitors will reversely modify the deviating pattern of the DNA methylation by interfering the enzymatic activity of the DNMTs.17,18 The current study aimed to investigate if there is a relationship between sequence variation in pap E , presence of Dam and susceptibility to antibiotics in UPEC.
Material and Methods
Bacterial Isolation
Sixty Five isolates of E. coli from urine specimens were obtained from hospitalized and out-patients suffering from signs and symptoms of UTI recruited to Department of Microbiology Lab,AL-Kadiymia Teaching Hospital, Baghdad, Iraq, from March 2018 to October 2018. Also, 2 E. coli isolates from wound infections and 2 E. coli isolates from chest fluid were obtained. These isolates were cultured on MacConkey agar and identified using API20E Epi 20 system and/or VITEK2 Vitic system. Information such as age, gender, antibiotic susceptibility of included patients were obtained from consent form of each patient.
Antimicrobial Susceptibility Testing
The antibiotic susceptibility test of included isolates were done using disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.19 The following antibiotics were included: amoxicillin-clavulanic acid (AMC), ciprofloxacin (CIP), gentamicin (GE), nitrofurantoin (NIT), trimethoprim-sulfamethoxazole (SXT), cefpodoxime (CPD),cephalothin (CLT), tetracycline (TE), rifampin (RP)and ticarcillin(TIM).The definitions of multidrug resistant (MDR),extended drug resistant (XDR) and pan drug resistant (PDR) isolates were as per standardized by European Centre for Disease Control (ECDC) and Centre for Disease Control & Prevention (CDC).20
Molecular Identification of E. Coli
Genomic DNA was extracted from E. coli isolates using WIZARD Genomic DNA Extraction Kit following manufacturer instructions (Promega,USA). Concentration and purity of extracted DNA of each sample were measured using Nanodrop (AcT Gene NAS-99, USA). Primers specific for the lacY (lactose permease gene) and phoA (bacterial alkaline phosphatase gene) were used for molecular identification of E. coli following Das Mitra et al., 2015with modifications.21
The reaction was prepared in 25μl total volume. The concentration of primer was 0.15μM for lacY, and 0.6μM for phoA(Alpha DNA,Canada),1X PCR buffer (Promega,USA), 1.5 U of Taq DNA polymerase(Promega,USA), 200 mM dNTPs(Promega,USA). Then, DNA(100 ng) was added. The cycling conditions were as followings: 94 ºC for 5 min followed by 35 cycles of 94 ºC for 30 sec, 52ºC for 1min and 72 ºC for 1 min and final extension at 72oC for 10 min using Thermal Cycler (Eppendorf master cycler,Germany). The amplified products were analyzed using agarose gel 1.5%. Extracted DNA from Pseudomonas aeroginosa identified using VITEK2 VITIC system and molecularly from previous study, 22 and extracted DNA from Klebsiella pneumoniae (identified using VITIC system in this study) were used as negative control to evaluate primer specificity. Two bands with molecular size 289bp and 468bp will be seen if the DNA was from E. coli. The appearance of amplicon with 468bp only or no amplified products will be refereed to bacteria other than E. coli.
Detection of pap E
The presence of pap E in UPEC isolates was detected using conventional PCR.23 Master mix was prepared in 25μL total volume containing 1X PCR buffer,10 pmol of forward and reverse primers, 200μM of dNTP, 1.25U of Taq DNA polymerase. Then, 100ng DNA was added. Two extracted DNA samples from each of E. coli isolated from wound infection and chest fluid, respectively, were used as negative control for the presence of pap E. PCR conditions were as following: 94oC for 2min followed by 35 cycles of 94°C for 1 min, 52°C for 1.5min, 72oC for 3min and final extension at 72oC for 10 minutes using Thermal Cycler (Eppendorf master cycler,Germany). PCR products were electrophoresed on agarose gels1.5%. Appearance of amplicon with molecular size 336bp referred to presence of pap E.
DNA Sequencing of pap E
The amplicons of pap E from 21 selected strains of UPEC were send for Sanger sequencing using automated DNA sequencer ABI 3730XL (Macrogen Corporation, Korea). The results were analyzed using Genious software. The sequences of each fragment were trimmed to a uniform length that corresponded with the region used to identify the target gene. Sequences were compared with standard strain MH455215 using online BLAST software (http: //www.ncbi.nlm.nih.gov/BLAST/). Partial sequencing tree of fimbrial adaptor papE and fimbrial adaptor papF of 13 E. coli isolates was constructed.
Nucleotide Sequence Accession numbers
The partial sequencing alignment of pap E of 5 included strains were compared with the sequences of previously published strains from DNA GenBank sequences using Basic Local Alignment Search Tool (BLAST) search and deposited in GeneBank under accession numbers: LC479519.1 of strain MR-1,LC479520.1 of strain MR-2, LC479521.1 of strain MR-14, LC479522.1 of strain MR-18 and LC 479523.1 of strain MR-21.
Detection of DNA Adenine Methyl-Transferase Gene in UPEC Isolates
Extracted DNA from 26 UPEC was screened for Dam using conventional PCR.24 PCR condition was modified as the following: 95oC for 5min followed by 35 cycles of 95°C for 1 min,55°C for 1.5min, 72oC for 1.5min and final extension of 72oC for 10 min using Thermal Cycler (Eppendorf master cycler,Germany). Products of PCR were electrophoresed on agarose gels0.8%. Appearance of amplicon with molecular size of 1071bp referred to presence of Dam.
Results
Patients Demography
The mean age of included patients was 38.11 ±22.02 ranging from (1-78) years with median 42.5 (36.5) years. Male 24/64 (37.5 %) to female 40/64 (62.5 %) ratio 1:1.7.
Prevalence of pap E among UPEC Isolates
Of the obtained 65 isolates, 64(98.4%) were approved to be E. coli depending on molecular identification of lacY and phoA using PCR. Detection of papE in E. coli strains using PCR revealed that 26/64(42.6%) were UPEC (Fig.1).Prevalence of UPEC was showed to be the highest among the female patients within age group (10-20) years and (50-60) years (Table1).
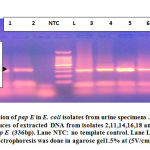 |
Figure 1: Detection of pap E in E. coli isolates from urine specimens. |
Table 1: Age distribution of patients infected with E. coli positive to papE UPEC.
| Age group (year) | Female | |
| Male | ||
| 10-Jan | 1 | – |
| 20-Oct | – | 5 |
| 20-30 | – | 1 |
| 30-40 | 2 | 2 |
| 40-50 | 1 | 3 |
| 50-60 | – | 5 |
| 60 and more | 3 | 3 |
| Total | 7 | 19 |
Analysis of pap E Sequencing in Studied Strains
PCR amplified products of papE from 21strains were partially sequenced, only 13 strains gave successful sequencing analysis of target gene which reviled three clades labeled as A,B and C (Fig. 2). The correlation between studied strains according analysis of genetic distance based on partial sequencing of pap E showed that strain 17(clade A) have 100% similarity with standard strain, while clade B, which included strains (1,2,5,14,18,21) and clade C which included strains (4,7,8,10,12,13.16) have (99%) similarity with standard strain.
From clade B, strains 1, 2 and 21 considered as sister group. Also, strains 5, 14 and18 considered as sister group. From clade C, strains 12, 4, 7, 8,10,13,16 have similarity of 100%. The main clusters that were similar were conserved.
 |
Figure 2: Partial sequencing tree of pap E . |
Analysis of nucleotide sequence changes from partial sequencing of pap E shown that UPEC included in clade B displayed the most nucleotide sequence changes as substitution result in synonymous and non-synonymous mutation (Table 2).
Prevalence of Dam among UPEC Isolates
Dam was detected in 11/64 (17.1%)E. coli isolates (Fig. 3). The association between the presence of pap E and Dam in studied isolates shown that 10/64(15.6%) isolates contain both genes (Table 3).
Table 2: The sequencing analysis of partial sequencing tree of pap E
| Clade type of pap E | Strain no. | Nucleotide change | Amino-acid change | Type of nucleotides change |
| A | 17 | N | N | N |
| 1 | GTC/GTT | Val-Val | Substitution (synonymous) | |
| TGG/TAA | Val-Val | Substitution(synonymous) | ||
| 2 | GTC/GTT | Val-Val | Substitution(synonymous) | |
| B | TGG/TAT | Trp-Tyr | Substitution(non-synonymous) | |
| 5 | GAA/GAG | Glu-Glu | Substitution(synonymous) | |
| ATC/GTA | Lie-Val | Substitution (non-synonymous) | ||
| 14 | GAA/GAG | Glu-Glu | Substitution(synonymous) | |
| ATC/GTA | Lie-Val | Substitution(non-synonymous) | ||
| TTC | Phe | Insertion-frame shift | ||
| 18 | GCT/ATT | Ala-Lie | Substitution(non-synonymous) | |
| GTC/GTT | Val-Val | Substitution(synonymous) | ||
| ATC/GTC | Lie-Val | Substitution(non-synonymous) | ||
| 21 | GTC/GTT | Val-Val | Substitution(synonymous) | |
| TTC | Phe | Insertion-frame shift | ||
| 4 | N | N | N | |
| 7 | N | N | N | |
| C | 8 | N | N | N |
| 10 | N | N | N | |
| 12 | N | N | N | |
| 13 | N | N | N | |
| 16 | N | N | N |
N: no change in nucleotides, no change in amino acid; Val: valine ; Trp: tryptophan ; Tyr: tyrosine; Glu: glutamine; Lei: leucine ; Phe: phenylalanine ; Ala: alanine.
 |
Figure 3: Detection of Dam in E. coli isolates from urine specimens. |
Table 3: Frequency distribution and prevalence rate of pap E and Dam
| Characteristic | N=64 | (%) |
| Pap E -/ Dam – | 37 | 57.8 |
| Pap E +/ Dam – | 16 | 25 |
| Pap E -/ Dam + | 1 | 1.6 |
| Pap E +/ Dam + | 10 | 15.6 |
| Pap E+ | 26 | 40.6 |
| Dam + | 11 | 17.2 |
Antimicrobial Susceptibility Test
Of the UPEC isolates, 13/26 (50%) isolates were considered as MDR, that acquired non-susceptibility to at least one antibiotic in three or more antimicrobial categories (Table S1).
Association between pap E clades of UPEC and MDR
Studying the association between the pap E clades of UPEC isolates and MDR was shown that there was statically not significant (Table 4).
Table 4: UPEC pap E clads in association with MDR.
| Clad Type | Total | MDR + | MDR – | P | |
| A | 1 | 0 | 1 | 0.462 F
NS |
A versus others |
| B | 6 | 4 | 2 | 0.592 F
NS |
B versus others |
| C | 6 | 3 | 3 | 1.000 F
NS |
C versus others |
| Total | 13 | 7 | 6 | — | — |
F: Fischer exact test; NS: not significant at p ≤ 0.05
Correlation between Antibiotic Susceptibility, MDR and the Existence of pap E and Dam
The correlation between the presence of papE and /or Dam and antibiotic susceptibility reviled that there was a statistically significant correlation between resistance to tetracycline and rifampin and the presence of both papE and Dam, a statistically significant correlation between resistance to tetracycline and the presence of pap E and a high statistically significant correlation between resistance to tetracycline and the presence of Dam (Table S2). The study of MDR risk in association with the presence of pap E and Dam in UPEC revealed that Dam could be considered as etiological factored to developing MDR (Table 5).
Table 5: Multidrug resistance risk in association with pap E and Dam
| Gene | MDR +
n = 28 |
MDR –
n = 36 |
P | OR | 95 % CI | EF | ||
| n | % | n | % | |||||
| pap E | 13 | 46.4 | 13 | 36.1 | 0.404 C
NS |
1.53 | 0.56 -4.20 | 0.17 |
| Dam | 8 | 28.6 | 3 | 8.3 | 0.047 F
S |
4.40 | 1.04 -18.54 | 0.56 |
n: number of cases; OR: Odds ratio; CI: confidence interval; EF: etiologic fraction; C: Chi-square test; F: Fischer exact test; NS: not significant at P ≤ 0.05
Discussion
Colonization of E. coli strains within the urinary tract and occurrence of UTI begin with binding of bacteria to epithelial surface.25,26In the current study, 26/64(42.6%) of E.coli were confirmed to be UPEC according to molecular detection of papE. There is a diversity in the frequency of pap gene among UPEC strains across the globe and within the same geographical region. Iraqi studies at 2015 and 2017 referred to that pap was detected in 58/112 (51.785%) E. coli and 31/43(72.09%) E. coli, respectively, isolated from urine of patients with UTI.27,28 Iranian studies were revealed that there were variations in pap gene prevalence in UPEC isolated from different locations including 83.63%, 16.6% and 20.5%.29,30,31 Indian study at 2018 found that the prevalence of pap was 72/350(20.5%) isolates.26 This variation result from UPEC use a variety of adhesions to attach to the bladder urothelium if the usual adhesions are not expressed. However, there is always the possibility of occurring mutations in pap result in missing of its detection. A positive PCR product confirm the detection of the gene, but a negative result does not insure absence of the gene.
Increased antibiotic resistance in bacteria causing UTI has complicated the treatment of such infections. In the current study, 13/26(50%) UPEC isolates were MDR.Iraqi study referred to that of the 62 E. coli isolated from urine, (69.4%) were showed MDR32.Iraqi stud was found that 44% of UPEC were resist to ciprofloxacin and these isolates have mutation in gyrA that identified using sequencing of this gene.33 Other Iraqi study at 2018 found that of 42 UPEC, 37(88.09%) were found to be MDR34. Iranian study at 2015 found that 111/150(74%) of UPEC strains showed MDR phenotype.31
In the current study, the etiological factor of Dam in MDR isolates was 0.56. That referred to the possibility of considered the presents of Dam as one of predisposing factor for resistance. A study was done to determine the effect of the bacterial epigenome in antibiotic stress survival to describe genomic methylation kinetics using single-molecule real-time sequencing and they find that without adenine methylation at GATC sites, E. coli growth under antibiotic stress is extremely reduced. The explanation for the role of Dam in antibiotic resistance is that during drug stress, the adenine methylome persist stable but without GATC methylation, methyl-dependent mismatch repair is repressed and that cause toxic DNA breaks in bacteria. In drug-resistant and pathogenic E. coli strains, Dam insufficiency reduce responsiveness to antibiotics such as β-lactam and quinolone classes.35 A study referred to that there was an 8-fold decrease in resistance for amoxicillin/clavulanic acid, gentamicin and trimethoprim/sulfamethoxazole and 4-fold decrease in resistance for ciprofloxacin in Dam deleted mutated UPEC strains.36
Current studies are focus on the detection of Dam,17,37 but some E. coli strains could be have DNA cytosine methyltransferase (Dcm) that methylates the second cytosine in the sequence 5′CCWGG 3′.13,38 Also, it was referred to that Dam could be recognized and methylate cytosine in sequences other than GATC.39
Conclusion
The study of multi-drug resistance(MDR) risk in association with the presence of pap E and Dam in UPEC revealed that Dam may be considered as etiological factored to developing MDR. Dam should be taken in consideration as one mechanism of MDR prediction development in UPEC. Farther studies included a large sample size required to sport the role of pap and Dam in responsiveness to antibiotic.
Supplementary Material
Table S1: Antimicrobial susceptibility test of UPEC.
| Patient No. | Age | Gender | Pap / Dam | Antibiotics | Resistance type | |||||||||||
| (Y) | GE | NIT | SXT | CPD | AMC | CLT | TIM | RP | TE | CIP | ||||||
| 1 | 48 | M | +/+ | S | S | R | R | R | R | R | S | R | R | MDR | ||
| 2 | 35 | F | +/+ | S | S | R | R | R | R | R | S | R | S | MDR | ||
| 3 | 78 | M | +/ – | S | S | R | R | R | R | S | S | S | S | – | ||
| 4 | 54 | F | +/ – | S | S | R | R | R | R | S | R | S | S | MDR | ||
| 11 | 14 | F | +/ – | S | S | S | S | S | S | R | S | R | S | – | ||
| 12 | 23 | F | +/ – | S | S | S | S | S | S | R | S | R | S | – | ||
| 14 | 43 | F | +/ – | S | S | S | S | S | S | R | S | R | S | – | ||
| 16 | 65 | F | +/ – | S | S | S | S | S | S | R | S | R | S | – | ||
| 18 | 50 | M | +/ – | S | S | S | S | S | S | R | S | R | S | – | ||
| 19 | 33 | M | +/+ | S | S | S | S | S | S | R | S | R | S | – | ||
| 20 | 46 | M | -/+ | S | S | S | S | S | S | R | S | R | S | – | ||
| 21 | 9 | F | +/+ | R | S | S | S | S | S | R | R | R | S | MDR | ||
| 22 | 64 | M | +/+ | S | S | S | S | S | S | S | R | S | R | MDR | ||
| 23 | 30 | F | +/+ | S | S | R | R | R | R | R | R | R | R | MDR | ||
| 24 | 36 | M | +/+ | S | S | R | R | R | R | R | R | S | R | MDR | ||
| 26 | 1 | M | +/ – | S | S | S | S | S | S | S | R | S | S | – | ||
| 31 | 76 | M | +/- | S | S | S | S | S | S | R | S | S | R | – | ||
| 32 | 6 | F | +/+ | S | S | S | S | S | S | R | S | S | R | – | ||
| 34 | 11 | F | +/ – | S | S | S | S | S | S | R | R | R | S | MDR | ||
| 35 | 10 | F | +/ – | R | R | S | S | S | S | R | R | R | S | MDR | ||
| 36 | 44 | F | +/+ | S | R | R | R | R | R | R | R | R | R | MDR | ||
| 48 | 60 | F | +/+ | R | S | S | S | S | S | R | R | R | R | MDR | ||
| 53 | 56 | F | +/- | S | S | S | S | S | S | S | S | S | R | – | ||
| 57 | 48 | F | +/- | S | S | R | S | S | S | S | S | S | R | – | ||
| 58 | 62 | F | +/- | S | S | S | S | S | S | S | S | S | R | – | ||
| 61 | 57 | F | +/- | S | S | R | R | R | R | R | S | S | R | MDR | ||
| 63 | 55 | F | +/- | S | S | R | R | R | R | R | R | S | R | MDR | ||
M: male; F:female;S: sensitive; R: resist; MDR: multi-drug resistance; AMC: amoxicillin-clavulanic acid, CIP: ciprofloxacin, GE: gentamicin, NIT:nitrofurantoin, SXT: trimethoprim-sulfamethoxazole (SXT),CPD: cefpodoxime, CLT:cephalothin, TE:tetracycline,RP: rifampin and TIM:ticarcillin.
Table S2: Correlation between papE and/or Dam and antibiotic susceptibility.
| Variable | pap E | Dam | ||
| r | p | r | p | |
| Gentamycin | 0.115 | 0.366 | 0.176 | 0.164 |
| Nitrofurantion | -0.048 | 0.708 | -0.004 | 0.972 |
| Trimethoprim. | -0.062 | 0.624 | 0.030 | 0.813 |
| Cefpodoxime. | 0.004 | 0.974 | 0.106 | 0.403 |
| Amoxyclave | 0.032 | 0.803 | 0.123 | 0.334 |
| Cephalothin | 0.004 | 0.974 | 0.106 | 0.403 |
| Ticarcillin | 0.281 | 0.025* | 0.318 | 0.010* |
| Rifampien | 0.261 | 0.037* | 0.268 | 0.032* |
| Tetracycline | 0.279 | 0.025* | 0.331 | 0.007** |
| Ciprofloxacin | -0.078 | 0.541 | 0.082 | 0.520 |
| MDR | 0.104 | 0.412 | 0.266 | 0.034* |
*Significant at P ≤ 0.05; **highly significant at P ≤ 0.01.
Acknowledgments
Not applicable.
Ethics Statement
Not applicable
Conflict of Interest
The author declare that there is no conflict of interest.
Funding Source
There is not funding source
References
- Zalmanovici T. A., Green H., Paul M., Yaphe J. and Leibovici L. Antimicrobial agents for treating uncomplicated urinary tract infection in women. Cochrane Database Syst. Rev.2010;10:CD007182. doi:10.1002/14651858
- Maroncle N.M., Sivick K.E., Brady R., Stokes F.E. and Mobley H.L.T. Protease activity, secretion, cell entry, cytotoxicity and cellular targets of secreted autotransporter toxin of uropathogenic Escherichia coli. Infect. Immun. 2006;74:6124–34.. doi:10.1128/IAI.01086-06.
- Li K., Zhou W., Hong Y., Sacks S.H. and Sheerin N.S. Synergy between type I fimbriae expression and C3 opsonization increases internalisation of E. coli by human tubular epithelial cells. BMC Microbiol, 2009;9:64. doi:10.1186/1471-2180-9-64.
- Melican K., Sandoval R.M., Kader A., Josefsson L., Tanner G.A., Molitons B.A. Uropathogenic Escherichia coli P and type I fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog. 2011;7:e100298. doi:10.1371/journal.ppat.100129821.
- Ali Mohammed H., Falah Hasan S. and Ausama Ayoub J. Urinary Tract Infection Prevalence and Antibiotic Resistance A Retrospective Study in Basra Governorate, Iraq. AJPS. 2014;14:129-135.
- Abdulghani M. Alsamarai, Ibrahim Abdul-Rahman L. and Mohamed Mohsen A.A. Urinary tract infection in Iraq: Evaluation of early detection methods and etiology. World J. Pharmacy and Pharmaceutical. 2016; 5: 181-194. 10.20959/wjpps20167-7196.
- Karzan Abdulmuhsin M. , Zirak F. A. and Bayar H. M.. Determination the site of antibiotic resistance genes in Escherichia coli isolated From Urinary Tract Infection. Kurdistan J. Applied Research.2018;3: 2411-7684. DOI: 10.24017/science.2018.3.2.
- Munaf A. Aaboda1and Mohammed R. Al-Notazy. Antibiotics susceptibility profile of Escherichia coli isolated from patients with urinary tract infection in Misan, Iraq. J. Pharm. Sci. & Res., 2018; 10:2858-2861.
- Usein C.R., Damian M., Tatu-Chitoiu D., Capusa C., Fagaras R., Tudorache D., Nica M. and Le Bouguenec C. Prevalence of virulence genes in Escherichia coli strains isolated from Romanian adult urinary tract infection cases. J. Cell Mol. Med. 2001;5:303-10.
- Yun K.W., Kim H.Y., Park H.K., Kim W. and Lim I.S. Virulence factors of uropathogenic Escherichia coli of urinary tract infections and asymptomatic bacteriuria in children. J. Microbiol Immunol. Infect.2014;47:455-61. doi: 10.1016/j.jmii.2013.07.010.
- Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chem Bio Chem,2002;3: 274–293.
- Zemach A., McDaniel I.E., Silva P. and Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science.2010;328:916–919.
- Marinus M.G. and Casadesus J. Roles of DNA adenine methylation in host pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 2009;33:488–503.
- Adhikari S. and Curtis P.D. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol. Rev. 2016;40: 575-591. doi: 10.1093/femsre/fuw023.
- van der Woude M.W., Kaltenbach L.S. and Low D.A. Leucine-responsive regulatory protein plays dual roles as both an activator and a repressor of the Escherichia coli pap fimbrial operon. Mol. Microbiol.1995;17: 303–312.
- Makrina T., Scott A. B., Nicola H. and David L. G. Regulatory interplay between pap operons in uropathogenic Escherichia coli. Mol. Microbiol. 67: 996–1011. doi:10.1111/j.1365-2958.2007.06098.x
- Umairah N. M. O. and Suresh K. Bacterial DNA Adenine Methyltransferase as a Novel Drug Target for Antibiotics: Current Status and Future Drug Discovery Challenges. Internatio. J.Current Microbiol. Applied Sci.2019;8: 2494- 2504.
- Xu P., Hu G.,Luo C. and Z. Liang. DNA methyltransferase inhibitors : an updated patent review (2012-2015). Expert Opinion on Therapeutic Patent.2016;26:1017-30. doi: 10.1080/13543776.2016.1209488.
- Clinical and Laboratory Standards Institute, “Performance standards for antimicrobial susceptibility testing: 22nd informational supplement,”CLSI Document M100-S22, Clinical and Laboratory Standards Institute,Wayne, Pa, USA, 2012.
- Silpi B., Priyanka S. and Monali R. Multidrug Resistant and Extensively Drug Resistant Bacteria: A Study. Pathogens.2016;2016:5. http://dx.doi.org/10.1155/2016/4065603.
- Susweta D. M. , Wilfred A. D., Rituparna T., Nimita V. C.,Bhuvana M., Krithiga N., Bibek R. S. and Habibur R. Duplex PCR for specific detection of Escherichia coli and its differentiation from other Enterobacteriaceae. Indian J. Animal Sci.2015, 85 : 832-835.
- Jaafar Z.M., Dhahi M.A.R., Abd A.K. H. and Jaafar S.M. Molecular identification and antibiotics resistance genes profile of Pseudomonas aeruginosa isolated from Iraqi patients. African J. Microbiol. Res.2014,8(21):2183-2192.
- Yamamoto S., Terai A., Yuri K., Kurazono H., Takeda Y. and Yoshida O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol. Med. Microbiol.1995,12: 85-90.
- Stacy Ann-Marie S. and Paul D. B.. Eepigenetic influence of Dam Methylation on gene expression and attachment in Uropathogenic Escherichia coli. Frontiers in Public Health.2016;4:1-17. doi: 10.3389/fpubh.2016.00131
- Dale A.P. and Woodford N. Extra-intestinal pathogenic Escherichia coli (ExPEC): disease, carriage and clones. J. 2015, 71:615–626.doi:10.1016/j.jinf.2015.09.009.
- Sabitha B., Vimal K.K. and Rishiyur K.G. Determination of Adhesion Encoding Genes of Uropathogenic Escherichia coli. Avicenna J. Clin. Microbiol. 2018,5:20-26. doi:10.15171/ajcmi.
- Esam. G. M. S., Mohammed. I. N. and Maarib. N. R. Rapid Detection of Uropathogenic Escherichia coli virulence factoros in Iraqi patients by multiplex polymerase chain reaction. Word J. Pharma. Res.2015,4:507-515.
- Saba N. Abdul-Ghaffar and Rasmia A. Abu-Risha. Virulence Genes Profile of Escherichia coli Isolated from Urinary Catheterized and Non-Catheterized Patients. Iraqi J. Sci. 2017, 58: 820-835.doi:10.24996.ijs.2017.58.2B.6.
- Rahdar M., Rashki A. and Miri H. Comparison of the Common Adhesin Coding Operons Distribution in Uropathogenic and Phylogenetic Group B2 and A Escherichia coli Isolates. Avicenna J. Clin. Microbiol. Infect.2014;1:e22981. doi:10.17795/ajcmi-22981.
- Mohajeri P., Khademi H., Ebrahimi R., Farahani A.and Rezaei M. Frequency distribution of virulence factors in uropathogenic Escherichia coli isolated from Kermanshah in 2011-2012. Int. J. Appl. Basic Med. Res. 2014;4(2):111-6. doi: 10.4103/2229-516x.136794.
- Neamati F., Firoozeh F., Saffari M. and Zibaei M. Virulence Genes and Antimicrobial Resistance Pattern in Uropathogenic Escherichia coli Isolated From Hospitalized Patients in Kashan,Iran. Jundishapur J. Microbiol. 2015;8:e17514. doi: 10.5812/jjm.17514.
- Mohanad M. A. Incidence of Multi-Drug Resistant Escherichia coli Isolates from Blood and Urine in Kerbala, Iraq. Kerbala University .2014;12:222-227.
- Ali A. M. and Harithjabbar F. Al-Mathkhury. Prevalence of Ciprofloxacin resistant coli in urinary tract infections. IOSR J. Pharma. Biolog. Sci. 2018;13:25-28. doi: 10.9790/3008-1304012528.
- Huda H. Al-Hasnawy, Mohammad R. Judi and Hiba J. Hamza. The Dissemination of Multidrug Resistance (MDR) and Extensively Drug Resistant (XDR) among Uropathogenic coli (UPEC) Isolates from Urinary Tract Infection Patients in Babylon Province, Iraq. Baghdad Sci. J.2019;16:986- 992. doi:http://dx.doi.org/10.21123/bsj.2019.16.4(Suppl.).0986.
- Nadia R. Cohen, Christian A. Ross, Saloni J., Rebecca S. Shapiro, Arnaud G, Peter B., Hu L. and James J. Collins. A role for the bacterial GATC methylome in antibiotic stress survival. Genet., 2016; 48: 581–586. doi:10.1038/ng.3530.
- Paul D. B. Dam methylation has profound effect on antibiotic resistance in uropathogenic Escherichia coli. J. Med. Microb. Diagn. 2017;6:3.doi: 10.4172/2161-0703-C1-012.
- David R., Hervé N., María A. Sánchez-Romero, Ignacio C.,Dan I. A. and Josep C. A. Portable epigenetic switch for bistable gene expression in bacteria. Scientific Rep.2019; 9:11261. https://doi.org/10.1038/s41598-019-47650-2
- Kevin T. Militello, Robert D. Simon, Mehr Q., Robert M., Michelle L. Van H., Stacy M. Hennick, Sangeeta K. Jayakar and Sarah P. Conservation of Dcm-mediated Cytosine DNA Methylation in Escherichia coli. FEMS Microbiol. Lett. 2012; 328:78–85. doi:10.1111/j.1574-6968.2011.02482.x.
- John R. Horton, Xing Zhang, Robert M. Blumenthal and Xiaodong Cheng. Structures of Escherichia coli DNA adenine methyltransferase (Dam) in complex with a non-GATC sequence: potential implications for methylation-independent transcriptional repression. Nucleic Acids Res., 2015; 43: 4296–4308. doi: 10.1093/nar/gkv251.









