Haris Nur Mustofa1 , Hesti Lina Wiraswati1, 2*
, Hesti Lina Wiraswati1, 2* , Savira Ekawardhani1,2
, Savira Ekawardhani1,2 , Pandji Irani Fianza3
, Pandji Irani Fianza3 and Sarasati Windria4
and Sarasati Windria4
1Oncology and Stem Cell Working Group, Faculty of Medicine, Universitas Padjadjaran, 45363, Jatinangor, Jawa Barat, Indonesia
2Parasitology Division, Department of Biomedical Science, Faculty of Medicine, Universitas Padjadjaran, 45363, Jatinangor, Jawa Barat, Indonesia
3Department of Internal Medicine, Faculty of Medicine, Universitas Padjadjaran, 45363, Jatinangor, Jawa Barat, Indonesia
4Infection Study Centre, Faculty of Medicine, Universitas Padjadjaran, 45363, Jatinangor, Jawa Barat, Indonesia
Corresponding Author E-mail : hesti.lina@unpad.ac.id
DOI : https://dx.doi.org/10.13005/bpj/1969
Abstract
One of the known causes of cancer is imbalance production between reactive oxygen species (ROS) and antioxidant defense within the cell. Under oxidative stress conditions, excessive ROS production ultimately induces cell death via apoptosis or necrosis. Moreover cancer cells use glycolysis for energy production. Glycolysis inhibition will lead to cancer cell proliferation disruption. Quinones and saponins are chemical compounds that have anticancer properties. Saponin is one of the plant’s metabolites, which also found in Indonesian medicinal plants. Preliminary studies in our lab showed that there were some species of medicinal plants in Indonesia that contained saponin, in which saponin known to have a good effect on inhibiting cancer cells. This study was aimed to know the anticancer activities from the compound that contains saponin and quinone as its active substance through the oxidative stress induction and glycolysis inhibition mechanism using in silico method. This research started by choosing the protein crystal structure of cytochrome p450 reductase (CYP450R) enzyme and pyruvate kinase M2 (PKM2) enzyme, then preparation of the protein with the help of Chimera 1.14.rc software, preparation of the ligands which belongs to cytotoxic molecule and optimization of their structure using Marvin Sketch software, and validation of molecular docking and docking process of the testing ligands on CYP450R and PKM2 enzymes using AutoDock Vina software. The results showed that testing ligands had affinity energy and good interaction with CYP450R and PKM2 enzymes, especially diosgenin. Testing ligands tended to interact with CYP450R rather than PKM2. Molecular interaction between testing ligands with the enzymes may provoke excessive ROS production and inhibit the glycolysis process in cancer cells. Compared to glycolysis inhibition, testing ligands had greater capacity in causing oxidative stress. Perhaps this study will motivate others for discovered the potential activity of medicinal plants in Indonesia.
Keywords
Anticancer; Glycolysis; Molecular Docking; Oxidative Stress; Quinone; Saponin
Download this article as:| Copy the following to cite this article: Mustofa H. N, Wiraswati H. L, Ekawardhani S, Fianza P. I, Windria S. Anticancer Activities of Saponins and Quinones Group through Oxidative Stress and Glycolysis Inhibition via in Silico Studies. Biomed Pharmacol J 2020;13(2). |
| Copy the following to cite this URL: Mustofa H. N, Wiraswati H. L, Ekawardhani S, Fianza P. I, Windria S. Anticancer Activities of Saponins and Quinones Group through Oxidative Stress and Glycolysis Inhibition via in Silico Studies. Biomed Pharmacol J 2020;13(2). Available from: https://bit.ly/2UdKdR3 |
Introduction
Cancer is one of the biggest causes of death in the world with an estimated 9.6 million deaths in 2018. According to GLOBOCAN data (Global Cancer Incidence, Mortality and Prevalence) the number of cancer cases that occurred in 2018 was 18.1 million cases in worldwide. Lung, colorectal, gastric, and liver cancers contribute the highest number of deaths annually.1 While in Indonesia alone the prevalence of cancer in 2018 is 1.8 per mil.2 Cancer cells have different behaviors with normal cells, including the energy source that comes from the process of glycolysis (the Warburg effect), increased biosynthesis of macromolecules, and differences in oxidative stress levels due to the production of ROS which are higher than normal cells.3
Small amounts of ROS can have a beneficial effect on cells, support cell proliferation and enhance cell survival. But when ROS experiences a high increase, it will cause the effects of adverse oxidative stress that can trigger cell death. To fight oxidative stress, a cell uses antioxidants to prevent excessive accumulation of ROS.4 In cancer cells, protein translation and deviant cell metabolism will increase the amount of aberrant ROS. ROS extraordinary control from cancer cells and their mechanisms against ROS can prevent cancer cells from the deadly effects of oxidative stress, but also increase the likelihood that cells can undergo additional mutations mediated by ROS and stress responses that encourage tumorigenesis.4
Cancer drugs have been used to kill cancer cells, some of which are known to use ROS production pathways, for example doxorubicin, cisplatin, mitoxantrone, and adriamycin. 4,5,6 The stress agents of the quinone group, such as menadione and benzoquinone also have a role in cancer cell death through the mechanism of increasing ROS production.7 Increased production of ROS is a quinone mechanism against the survival ability of cancer cells. The increased production of ROS will then trigger the oxidative stress process due to an imbalance between the number of oxidants and antioxidants in the cell.8 Quinone’s ability to induce oxidative stress is caused by its capacity to form radical hydroquinone and semiquinone compounds that are assisted by reductase enzymes including CYP450R.9
Furthermore, semiquinone can be oxidized by oxygen molecules which will cause the appearance of radical superoxide. This process will further generate a redox cycle to produce ROS, specifically hydrogen peroxide and hydroxyl radicals. Oxidative stress arising from an increase in ROS production will encourage cell death by mediating DNA lesions which will activate the apoptotic mechanism through p53 protein and damage the mitochondrial membrane.9 The next process is the release of pro-apoptotic agents (cytochrome C and Apoptosis-Inducing Factor) or causes permanent damage to the macromolecular components of a cell.10
Cancer cells also make the transition of energy from the electron transport chain in mitochondria to glycolysis, more specifically aerobic glycolysis. The process of glycolysis itself is regulated by 10 enzymes. There are 3 main enzymes have an important role, namely the enzyme hexokinase, phosphofructokinase, and pyruvate kinase.11,12 Pyruvate kinase enzyme plays a role in catalyzing the reaction of phosphate group transfer from the phosphoenolpyruvate compound (PEP) to adenosine diphosphate (ADP) which will produce one pyruvate molecule and one adenosine triphosphate (ATP) molecule.13 This enzyme can be inhibited by lapachol compounds.14
In addition, there are diosgenin and cauloside A molecules, both of which are Saponin groups. Saponins themselves can be found in medicinal plants in Indonesia, such as Plumeria rubra and Anredera cordifolio. Diosgenin influences the balance of cell oxidation and can induce apoptosis.15 Cauloside A shows a strong effect on the production of proinflammatory cytokines.16 Based on existing sources, it is necessary to know the cytotoxic activity of saponins and quinones group as ROS inducing agents and glycolysis inhibitors in cancer cells based on their interaction with the CYP450R enzyme and PKM2 enzyme using in silico method.
Materials and Method
Materials
The materials in this study used the crystal structure of the CYP450R enzyme (pdb id: 1AMO) and the PKM2 enzyme (pdb id: 4G1N) which could be choosen via https://www.rcsb.org and the three-dimensional structure of the cytotoxic group of quinones molecule (doxorubicin, menadione, benzoquinone), lapachol and saponins (diosgenin and cauloside) drawn by the conformer structure and prepared using the help of Marvin Sketch software by looking at the reference structure of the conformer from https://www.pubchem.ncbi.mlm.nih.gov
Tools
One laptop that has Windows 7 128 bit specifications, Intel i5 – 7750 processor, and 4GB RAM. The software used was AutoDock Vina on Windows OS, Chimera 1.14.rc, YASARA, and Marvin Sketch.
Research Method
Preparation of the CYP450R and PKM2 Enzymes
Preparation of the CYP450R enzyme and PKM2 enzyme started with selecting the crystal structure of a protein that was still complex in shape with a native ligand. The next stage was the selection of a protein chain. The selected chain was a chain that had a native ligand and had 100% similarity with other chains. After getting selected chains, the two enzymes were separated from their water molecules, heteroatoms and native ligands, respectively using the Chimera 1.14.rc software to make room for the testing ligands that will bind to the same place as the native ligand and make sure that interaction only happened between these enzymes and the ligand. Hereafter adding charges and hydrogen was carried out for the final step of the protein preparation.
Validation of the Molecular Docking
Validation of the molecular docking method was done by docking back the native ligand on the target protein that had been removed from its native ligand by using the AutoDock Vina software. The docking method that had been made was said to be valid if the RMSD (Root Mean Square Deviation) value obtained is less than 2Å and contains contact residues and hydrogen bonds so that docking between testing ligands and the protein can be done. To obtain a valid method, protein and ligand preparation must be carried out appropriately and the grid box was arranged to 1Å resolution and must accommodate the native ligand and all contact residues that bind to native ligand so that an RMSD value of less than 2Å is obtained.17
Three Dimensional Structure Optimization and Preparation of the Cytotoxic Molecule
The conformer structure from all of the ligands (doxorubicin, menadione, benzoquinone, lapachol, diosgenin and cauloside A) was drawn and optimized using the help of Marvin Sketch software. Reference of the conformer structure from all of the ligands can be obtained at https://www.pubchem.ncbi.mlm.nih.gov. Structure optimization was done by calculating the dreiding energy field. Furthermore, like the receptor, the ligand also got additional charges and hydrogen for the final preparation.
Docking of Cytotoxic Molecules on CYP450R and PKM2 Enzymes
Control ligands (doxorubicin, menadione, and benzoquinone) and testing ligands (lapachol, diosgenin and cauloside A) that had been optimized were docking with prepared CYP450R enzyme. While the control ligand for docking with PKM2 enzyme was lapachol and the others acted as testing ligands. The docking process was done by using the AutoDock Vina software.18 The grid box used in this process was a validated grid box. The results of the analysis will show the molecules (ligands) with the nine best conformation and binding energy to bind to the target protein.
Data Analysis
Analysis of the docking results includes the affinity energy of the bond, the number and types of contact residues, and the number of hydrogen bonds. The value of affinity energy or bond energy indicates the strength of the bond between the testing ligands and the receptor. The lower or the more negative the value of the bond energy, the stronger the bond between the molecule (ligand) and the receptor. Interactions that occured between doxorubicin, menadione, benzoquinone, lapachol, diosgenin and cauloside A with CYP450R enzyme and PKM2 enzyme can also be seen from the type of bonds formed.
Result and Discussion
Preparation of the CYP450R and PKM2 Enzymes
CYP450R consists of 3 chains, namely A, B, and C. Native ligand from CYP450R enzyme, nicotinamide adenine dinucleotide phosphate (NAP) which has oxidative properties located in A and B chains of CYP450R enzymes. This study used A chain for CYP450R preparation because it had 100% similarity and identical with B chain. CYP450R that had been separated and its native ligand were shown in Figure 1.
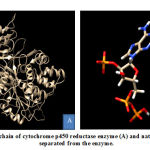 |
Figure 1: A chain of cytochrome p450 reductase enzyme (A) and native ligand (B) separated |
Meanwhile, the PKM2 enzyme composed of 4 chains, namely A, B, C, and D. Native ligand of PKM2 enzyme, N- (4 – {[4- (pyrazin-2-yl) piperazine-1-yl] carbonyl} phenyl) quinoline-8-sulfonamide (NZT) which act as activator is present in the A and D chains of PKM2 enzyme. This study used A chain for PKM2 enzyme preparation because of its 100% similarity and identical with D chain. PKM2 that had been separated and its native ligand were shown in Figure 2. CYP450R and PKM2 enzyme preparation were done by separating enzymes from its native ligands to provide space (pocket) which will be used for cytotoxic molecule docking with these enzymes. The output from this enzyme preparation process was enzyme structures without native ligands and its native ligands which were stored in the .pdb format.
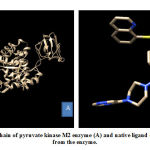 |
Figure 2: A chain of pyruvate kinase M2 enzyme (A) and native ligand (B) separated from the enzyme. |
Validation of the Molecular Docking
Validation of molecular docking method was carried out by redocking the prepared chain of CYP450R and PKM2 with its native ligand that had previously been separated using Chimera 1.14.rc software. The result and interaction figure of CYP450R docking validation were shown in Table 1 and Figure 3 and PKM2 docking validation were shown in Table 2 and Figure 4.
Table 1: Result of molecular docking validation on CYP450R enzyme
| Native ligand | Affinity energy (kcal/mol) | RMSD (Å) | Hydrogen bonds | Contact residues |
| NAP – reference | 10 (ARG298, THR535, SER596, ARG597, LYS602, TYR604, ASP639) | 7 (ARG298, THR535, SER596, ARG597, LYS602, TYR604, ASP639) | ||
| NAP – experimental | -10,3 | 0,4583 | 5 (ARG298, THR535, SER596, ARG597, LYS602) | 11 (ARG298, GLY534,THR535, CYS566, ARG567, SER596, ARG597, LYS602, TYR604, GLN606, ASP639) |
Table 2: Result of molecular docking validation on PKM2 enzyme
| Native ligand | Affinity energy
(kcal/mmol) |
RMSD (Å) | Hydrogen bonds | Contact residues |
| NZT – reference | 3 (LYS311, LEU353, TYR390) | 6 ( PHE26, MET30, LYS311, LEU353, TYR390, LEU394) | ||
| NZT- experimental | -9,1 | 0,3319 | 1 (TYR390) | 6 (PHE26, MET30, LEU353, TYR390, GLN393, LEU394) |
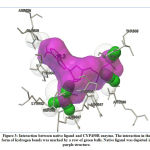 |
Figure 3: Interaction between native ligand and CYP450R enzyme. |
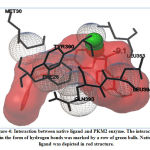 |
Figure 4: Interaction between native ligand and PKM2 enzyme. |
This docking method validation used RMSD value as a parameter which is the measurement of two poses with comparing the atomic position between the crystal structure and the structure that had been docked on the protein using the aid of the YASARA software. The method is considered to be valid if the RMSD value obtained is <2Å, so that the result produced from this validation shown that the docking method used was valid. The smaller the RMSD value produced indicates that the predicted ligand pose is getting better as it approaches or more similar to the native ligand.19
Three Dimensional Structure Optimization and Preparation of Cytotoxic Molecules
The three-dimensional structure of the testing ligands (doxorubicin, menadione, benzoquinone, lapachol, diosgenin, and cauloside A) made and optimized using Marvin Sketch software. The optimization results were shown in figure 5. This process started with drawing 3D conformer structures.
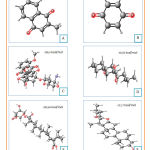 |
Figure 5: Three dimensional structures and dreiding energy field of menadione |
Furthermore, the structure was checked until the structure appeared to be valid and according to the template used. Then the calculation of dreiding energy field was carried out which will produce the 6 most stable conformations to choose from. The best dreiding energy field from each structure was shown in figure 5. Most stable conformation from each ligand saved in .pdb form. After that, ligand preparation carried out by adding charges and hydrogen to the cytotoxic molecules. Eventually, ligands were ready for the docking process.
Docking of Cytotoxic Molecules on CYP450R Enzyme and PKM2 Enzyme
Docking of cytotoxic molecules was carried out using AutoDock Vina software by applying validated grid box. The results obtained from the docking process between cytotoxic molecules with CYP450R and PKM2 enzymes in the form of affinity energy, contact residues, and hydrogen bonds formed. Results and visualization of docking interactions that occur between cytotoxic molecules and CYP450R and PKM2 enzymes were shown in Tables 3 and 4 as well as Figures 6 and 7.
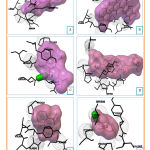 |
Figure 6: Visualization of interactions between Diosgenin |
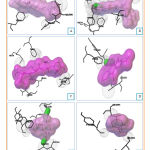 |
Figure 7: Visualization of interactions between Diosgenin |
The results obtained from the docking process between the testing ligands and the receptors (enzymes) in the form of affinity energy, hydrogen bonds and contact residues at the 9 best conformations. The affinity energy shows the bonds (interactions) between the testing ligands (diosgenin, doxorubicin, lapachol, cauloside A, menadione, and benzoquinone) and the enzymes (CYP450R and PKM2), the smaller the affinity energy obtained, the more stable the bond formed. The affinity energy between the testing ligands (diosgenin, doxorubicin, lapachol, cauloside A, menadione, and benzoquinone) and CYP450R enzyme and their comparison with the native ligand were shown in Table 3. Native ligand had the smallest affinity energy (most stable) meanwhile the testing ligands were found to have higher affinity energy, with diosgenin being the lowest one (most stable). It means that the testing ligands had to overcome the barrier (threshold) for replacing the native ligands’ role. The barrier may be concentration, temperature, et cetera.
Table 3: Docking results between cytotoxic molecules and CYP450R enzyme
| Ligand | Affinity energy (kcal/mol) | Hydrogen bonds | Contact residues |
| NAP – experimental | -10,3 | 5 | 11 (ARG298, GLY534,THR535, CYS566, ARG567, SER596, ARG597, LYS602, TYR604, GLN606, ASP639) |
| Diosgenin | -8,7 | – | 6 (PRO533, CYS566, ARG567, SER596, TYR604, VAL605) |
| Doxorubicin | -8,5 | 2 (ARG298) | 9 (ARG298, VAL474, GLY565, CYS566, TYR604, VAL605, ASP632, ARG634, SER678) |
| Lapachol | -7,5 | 1 (UNK0) | 5 (PRO533, TYR604, GLN606, ASN635, ASP639) |
| Cauloside A | -7,1 | – | 7 (ARG298, PRO533, GLY534, TYR604, ASP632, ASN635, MET636) |
| Menadione | -7,1 | – | 5 (PRO533, TYR604, VAL605, GLN606, ASN635) |
| Benzoquinone | -6,0 | 1 (SER596) | 5 (PRO533, CYS566, SER596, TYR604, VAL605) |
From the testing ligands which had been docked, doxorubicin, lapachol and benzoquinone formed the hydrogen bond with CYP450R enzyme. When they interact, all of the testing ligands had differences in number and types of contact residue. The number and types of contact residue from all of the testing ligands were shown in Table 3. TYR604 was the only contact residue that had been found in all of the testing ligands. It means that this residue may be a key for the enzyme to catalyze its reaction, and when this residue disappears the reaction may not be catalyzed.
CYP450R is a diflavoprotein enzyme whose function is providing electron transfer. This enzyme frequently found within the whole tissue and is concentrated highest in hepatocyte endoplasmic reticulum. CYP450R also has a role in xenobiotic biotransformation and endogenous molecule metabolism.20 Besides that, this enzyme also catalyzes the reduction of quinone to form semiquinone which will produce ROS.5 Testing ligands (diosgenin, cauloside A, and lapachol) were compared to the control (doxorubicin, benzoquinone, and menadione) which has evidence of ROS production through their interaction with CYP450R. The result showed that testing ligands also had good interaction with CYP450R as shown in Table 3.
Furthermore, lapachol which has evidence for providing an inhibitory effect on PKM2 was used as a control for testing ligands (diosgenin, doxorubicin, cauloside A, menadione, and benzoquinone) docking. The binding energy between testing ligands and PKM2 enzyme and their comparison with the native ligand were shown in Table 4. Native ligand had the smallest affinity energy (most stable) meanwhile the testing ligands were found to had higher affinity energy, with diosgenin being the lowest one (most stable). Again, it means that the testing ligands had to overcome the barrier for replacing the native ligands’ position. The barrier may be concentration, temperature, etc.
After the docking process on PKM2 enzyme had carried out, lapachol, doxorubicin, and menadione were found to form hydrogen bond. From the interaction between the testing ligands with the enzyme, it was clear that all of them had differences number and types of contact residue. The types and number of contact residue from all of the testing ligands were shown in Table 4. LEU353 was the only contact residue that had been found in all of the testing ligands. It means that this residue may be a key for the enzyme to catalyze its reaction, and when this residue disappears the reaction may not be catalyzed.
Table 4: Docking results between cytotoxic molecules and PKM2 enzyme
| Ligand | Affinity energy (kcal/mol) | Hydrogen bonds | Contact residues |
| NZT – experimental | -9,1 | 1 | 6 (PHE26, MET30, LEU353, TYR390, GLN393, LEU394) |
| Diosgenin | -7,5 | – | 5 (PHE26, LEU353, ASP354, GLU397, LEU398) |
| Doxorubicin | -7,2 | 1 (TYR390) | 8 (PHE26, MET30, GLY315, ASN318, LEU353, ALA388, TYR390, GLU397) |
| Cauloside A | -6,9 | – | 6 (PHE26, MET30, GLY315, ASN318, ASN350, LEU353) |
| Lapachol | -6,8 | 1 (TYR390) | 3 (PHE26, LEU353, TYR390) |
| Menadione | -6,2 | 2
(UNK0) |
6 (PHE26, LEU353, ASP354, TYR390, LEU394, GLU397) |
| Benzoquinone | -4,9 | – | 3 (LEU353, TYR390, LEU394) |
Pyruvate kinase is a metabolic enzyme whose function is to move the phosphate group from PEP to ADP in the process of glycolysis. This enzyme has several isoforms, namely PKM1, PKM2, PKL, and PKR found in mammalian cells and tissues.21 PKM2 enzymes are abundant on pulmonary, retinal, islet pancreatic cells and are also found in normal stem cells and embryonic cells. Regardless of the original tissue, the majority of cancer cells experience an increase in the expression of the PKM2 enzyme.22 PKM2 enzyme is a key enzyme in cancer cell metabolism, has a role in programming metabolism and is also directly involved in gene expression and cell cycle regulation.21,22 Testing ligands (diosgenin, doxorubicin, cauloside A, menadione, and benzoquinone) were compared to the control ligand (lapachol) which has evidence of glycolysis inhibition effect through its interaction with PKM2 enzyme. The result showed that testing ligands also had good interaction with PKM2 enzyme as shown in Table 4.
Based on the docking results, diosgenin, doxorubicin, lapachol, cauloside A, menadione, and benzoquinone may have potential anticancer activity because they had affinity energy to interact with CYP450R and PKM2 enzymes. According to the both interaction, testing ligands tented to interact with CYP450R rather than PKM2 as shown in Table 3 and Table 4. For example, diosgenin had better affinity energy with CYP450R compared to PKM2. It means that testing ligands had greater capacity to cause oxidative stress than glycolysis inhibition as anticancer agent.
Conclusion
In this present study, molecular interaction between all of the testing ligands and the receptors revealed that they had good interaction with CYP450R and PKM2 enzyme and may provoke excessive ROS production and inhibit the glycolysis process in cancer cells. The docking results unveiled that diosgenin shows the best affinity energy towards the CYP450R and followed by doxorubicin, lapachol, cauloside A, menadione, and benzoquinone, respectively. Testing ligands’ interactions with CYP450R will provide the lethal effect of ROS to make oxidative stress conditions and eventually cause the death of cancer cells. Docking results also acknowledged that diosgenin appearances the best affinity energy towards the PKM2 and followed by doxorubicin, cauloside A, lapachol, menadione, and benzoquinone, respectively. Their interactions with PKM2 will inhibit the glycolysis process to cause the reduction of ATP levels and ultimately restrain cancer cell proliferation. Testing ligands also showed greater capacity to cause oxidative stress than glycolysis inhibition as anticancer agent.
Acknowledgements
The author would like to humbly acknowledge the Internal Grant of Padjadjaran University.
Conflicts of Interest
All of the authors have no conflict interest upon the conduction of the experiment and content of the paper.
Funding Source
The author would like to humbly acknowledge the Internal Grant of Padjadjaran University.
References
- Bray F, Ferlay J, Soerjomataram I. Global Cancer Statistics 2018 : GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2018;68(6):394–424.
- Kementerian Kesehatan Badan Penelitian dan Pengembangan Kesehatan. Hasil Utama Riskesdas 2018. Kementeri Kesehat Badan Penelit dan Pengemb Kesehat. 2018;57.
- Panieri E, Santoro MM. ROS homeostasis and metabolism : a dangerous liason in cancer cells. Nature. 2016;7(6):2253–12.
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95.
- Wellington KW. Understanding cancer and the anticancer activities of naphthoquinones-a review. RSC Adv. 2015;5(26):20309–38.
- Bolton JL, Dunlap T. Formation and biological targets of quinones: Cytotoxic versus cytoprotective effects. Chem Res Toxicol. 2017;30(1):13–37.
- Wiraswati HL, Martoprawiro MA, Warganegara FM. Apoptosis Inducing Factor ( AIF ) Stabilizes Menadione- Conjugate Product in Programmed Cell Death. Int J PharmTech Res. 2017;10(4):237–45.
- Wiraswati HL, Hangen E, Sanz AB. Apoptosis inducing factor ( AIF ) mediates lethal redox stress induced by menadione. Impact Journals. 2016;7(47):76496–507.
- Saibu M, Sagar S, Green I, Ameer F, Meyer M. Evaluating the cytotoxic effects of novel quinone compounds. Anticancer Res. 2014;34(8):4077–86.
- Li XB, Gu JD, Zhou QH. Review of aerobic glycolysis and its key enzymes – new targets for lung cancer therapy. Thorac Cancer. 2015;6(1):17–24.
- Israelsen WJ, Vander Heiden MG. Pyruvate kinase: Function, regulation and role in cancer. Semin Cell Dev Biol. 2015;43:43–51.
- Phaniendra A, Jestadi DB, Periyasamy L. Free Radicals: Properties, Sources, Targets, and Their Implication in Various Diseases. Indian J Clin Biochem. 2015;30(1):11–26.
- Liberti M V., Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci. 2016;41(3):211–8.
- Babu MS, Mahanta S, Lakhter AJ, Hato T, Paul S, Naidu SR. Lapachol inhibits glycolysis in cancer cells by targeting pyruvate kinase M2. PLoS One. 2018;13(2):1–15.
- Atel KP, Adewar MG, Ahilyani VT, Kumar D, Atel P. A review on pharmacological and analytical aspects of diosgenin : a concise report. Nat Prod Bioprospect. 2012;2:46–52.
- Lee Y, Jung J, Ali Z, Khan IA, Oh S. Anti-Inflammatory Effect of Triterpene Saponins Isolated from Blue Cohosh ( Caulophyllum thalictroides ). Hindawi Publ Corp. 2012;2012:1–8.
- Hevener KE, Zhao W, Ball DM, Babaoglu K, Qi J, Stephen W, et al. Validation of molecular docking program. J Chem Inf Model. 2010;49(2):444–60.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading Oleg. J Comput Chem. 2011;31(2):455–61.
- Yadav DK, Khan F. Pharmacophore modeling , molecular docking , QSAR , and in silico ADMET studies of gallic acid derivatives for immunomodulatory activity. J Mol Model. 2012;18:2513–25.
- Gan L, Moltke LL Von, Trepanier LA, Harmatz JS, Greenblatt DJ, Court MH. Role of NADPH-Cytochrome P450 Reductase and Cytochrome- b 5 / NADH- b 5 Reductase in Variability of CYP3A Activity in Human Liver Microsomes. Am Soc Pharmacol Exp Ther. 2009;37(1):90–6.
- Yang W, Lu Z. Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett. 2013;339(2):153–8.
- Hsu M, Hung W. Pyruvate kinase M2 fuels multiple aspects of cancer cells : from cellular metabolism, transcriptional regulation to extracellular signaling. Molcular Cancer. 2018;1–9.








