Muhammad Nabil1,2* , Azman Seeni1
, Azman Seeni1 , Wan Ismahanisa Ismail2
, Wan Ismahanisa Ismail2 and Nurhidayah Ab. Rahim2
and Nurhidayah Ab. Rahim2
1Advanced Medical and Dental Institute, Universiti Sains Malaysia, 13200 Kepala Batas, Pulau Pinang, Malaysia.
2Faculty of Health Science, Universiti Teknologi MARA, Cawangan Pulau Pinang, Kampus Bertam, 13200 Kepala Batas, Pulau Pinang, Malaysia.
Corresponding Author E-mail: nabilfikrimail@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1755
Abstract
Cervical cancer is the third most common cancer affecting women worldwide. This occurs despite having precancerous screening and HPV vaccination implemented vigorously as a definitive intervention. Natural plant like Streblus asper has been discovered to offer great hope in treating and preventing cancers. In this study, we explored the potential of S.asper to inhibit the growth of cervical cancer cell line by using liquid chromatography mass spectrometry (LCMS). Upon analysis, seventy-six proteins that are common to both untreated and treated groups were identified. Of this, 14 proteins are found differentially expressed more than 2-fold changes. Based on past literature, we selected 7 proteins that are closely associated with treatment effects. These include Dermcidin, Keratin, type I cytoskeletal 9, Tropomyosin alpha-4 chain, Myristoylated alanine-rich C-kinase (MARCKS), Tumour protein D52, Folate receptor alpha, and Parathymosin. Pathway enrichment analysis by Reactome revealed 9 related pathways which include metabolism of protein, post-translational protein modification, signalling by Rho GTPases, signalling by NOTCH, cell cycle, cellular senescence, signalling by WNT, transcriptional regulation by TP53, and cellular responses to stress. These findings may improve our understanding on the related significant mechanism involving anti-cancer effects of S.asper on the cervical cancer cell line.
Keywords
Anticancer; Apoptosis; Cervical cancer; Natural product; Proteomics; Streblus asper
Download this article as:| Copy the following to cite this article: Nabil M, Nabil M, Seeni A, Ismail W. I, Rahim N. A. Proteomic Analysis of Anti-Cancer Effects of Streblus Asper Root Extract on HeLa Cancer Cells. Biomed Pharmacol J 2019;12(3). |
| Copy the following to cite this URL: Nabil M, Nabil M, Seeni A, Ismail W. I, Rahim N. A. Proteomic Analysis of Anti-Cancer Effects of Streblus Asper Root Extract on HeLa Cancer Cells. Biomed Pharmacol J 2019;12(3). Available from: http://biomedpharmajournal.org/?p=28501 |
Introduction
Cervical cancer has been ranked third most common cancer among females in the world.1 Cervical cancer arises from uncontrolled proliferation of cells at the cervix. In cervical malignancy, squamous cell carcinoma constitutes approximately 90% of the cases while another 10% is referred to as adenocarcinoma.2 Cervical cancer typically develops in a cervical transformational zone via Human Papillomavirus (HPV) transmission, viral persistence, progression of a clone of persistently infected cells to pre-cancer and invasion.3 As of today, numerous studies reveal various risk factors pertaining to cervical cancer development. These include Human Papillomavirus (HPV) infection, tobacco use, and long use of hormonal contraceptive pills.4,5,6,7 Among that, HPV infection has been identified to be the major risk factor that is responsible for up to 95% of cervical malignancies.8 The HPVs can be divided into two groups. There are low-risk group that are mostly associated with benign genital warts and the high-risk group that causes cervical cancer.9 HPV type 16 and 18 are the most commonly identified in invasive cervical cancer.10 In accordance with this, the HPV vaccination programme was introduced as a protective tool against this threat. This method has been widely accepted across the globe with approximately 70 countries establishing HPV vaccination as part of their national immunization programme.11 This trend has continued to be the practice for almost two decades. However, throughout the implementation, many weaknesses have been identified. HPV vaccination is considered expensive with the averaging cost of getting fully vaccinated from $1.49 to $18.94 per woman.12 This has been found hamper the efforts to widen the coverage of HPV vaccination especially in low- and middle-income countries.13 In addition, studies have found that parents with low knowledge on HPV vaccination are less likely to have their daughters vaccinated.14 This was driven by the fear of its possible adverse effects.15 Cervical cancer is generally controllably provided if the intervention is initiated at an early stage. Precancerous screening strategy is seen to offer great hope for the cervical cancer patient. A pre-cancerous screening which also known as Pap smear is a screening tool used to detect pre-cancerous changes like Cervical Intraepithelial Neoplasia (CIN) 1, CIN 2, and CIN 3. Women aged 30- to 49-year-old are recommended to undergo a screening process from time to time.16 This screening strategy has proven to provide effective protection for women above the age of 30 from cervical cancer mortality.17,18 In another study, the pap smear examination was observed to contribute to a 4% reduction in mortality.19 Despite its effectiveness, pap smear implementation has been observed to struggle against several challenges. These include lack of knowledge, failure in identification of eligible population, having difficulties in access that lead to demotivation of participation, having weaknesses in screening programme operation, insufficiency in monitoring and follow-up upon non-responders, and inadequacy of systematic monitoring of treatment.20 As for treatment, women who undergo radiotherapy and chemotherapy are unavoidably experienced adverse effects. Radiotherapy has been evidenced to cause urologic complications which include radiation cystitis, lower urinary tract dysfunction, stricture disease, fistula formation, and the development of second primary cancer.21 Radiotherapy is also often associated with acute side effects like erythema, desquamation, hair loss, mucositis, diarrhoea, pneumonitis, marrow ablation, nausea, and vomiting. While late or chronic side effects often result in fibrosis, necrosis, nerve damage, myelitis, telangiectasia, and stricture.22 Chemotherapy, on the other hand, is normally subjected to toxicities. It is observed to increase the risk of ovarian dysfunction in older age at the time of treatment.23 The anti-angiogenic agent which are commonly used in treating gynecologic malignancy is frequently found to produce various adverse events. These include hypertension, left ventricular dysfunction and congestive heart failure, acute vascular event, and bleeding tendencies.24 Based on these setbacks, an alternative approach to control its incidence and development is deemed to be imperative. In accordance with this, we suggested S.asper to be used as potential anticancer agent in this study. S.asper Lour is a family of Moraceae. It is a tree that normally grows indigenously in tropical countries like Sri Lanka, Malaysia, Thailand, the Philippines, and India.25 From root to leaf, S.asper extract and its constituents traditionally exploited to treat a diversity of maladies.26 Studies found that its extract can be used as anti-filarial, anti-fungal, anti-inflammatory, anti-microbial, anti-viral, anti-oxidant and anti-hyperglycemic, anti-diabetic, and anti-cancer.27,28,29,30,31,32,33,34,35,36,37 In addition, the role of S.asper extract as anti-cancer had been proved in cancers like osteosarcoma (HOS cells), tongue carcinoma (SCC-15 cells), mouse lymphocytic leukaemia (P388 cells), and human nasopharyngeal epidermoid carcinoma (KB cells).26,38,25 However, its possible effects against certain other types of cancer like cervical cancer remain uncertain. In this study, we unravelled the potential of S.asper to suppress cervical cancer cell line (HeLa cells) by studying the treatment effects at the proteomic level.
Materials and method
Plant extract
The Streblus asper plant was obtained from a nursery in Tasek Gelugor, Penang, Malaysia. The authenticity was later confirmed by Associate Professor Dr Md. Azman bin Pkm Seeni (Malaysian Institute of Pharmaceuticals and Neutraceuticals). S.asper roots were washed with distilled water and air-dried in an air-conditioned room for 2 weeks until it was completely dehydrated. The roots were ground into powders using Retstch SM 100 grinder. The ground powder was boiled with distilled water for 30 minutes and the outcome solution was filtered using 0.75 mm filter size. The filtrate was then freeze-dried. Upon usage, the freeze-dried powder’s weight was measured using analytical balance and diluted with double deionized distilled water according to the requirement.
Cell culture and treatment
All American Tissue Culture Collection cells used in this study were sub-cultured from ATCC with Catalog No. CCL-2 TM. This cervix adenocarcinoma cell line, HeLa cells (ATCC Catalog No.CCL-2TM) was bought from ATCC, Manasus, VA, USA. HeLa cells were maintained in DMEM supplemented with 10% FBS, 1% sodium pyruvate and 1% penicillin-streptomycin bought from Life Technologies, USA. Cells were incubated in 37°C humidified CO2 incubator, with 5% CO2 and 95% of air. The medium was replaced every 72 hours until it reached 80%-90% of cell confluency before sub-culturing was done. Three to ten cell passages were used in the experiment. As for protein analysis, S.asper treatment was performed on HeLa cells using the half maximal inhibitory concentration (IC50) dose (0.25 mg/ml) obtained from our previous study.
Protein digestion
The cell lysates prepared were undergone protein digestion. Prior to that, acetone precipitation of protein was done to eliminate substances that could interfere with LCMS application. The supernatant of prepared cell lysates was removed, and 80% cold acetone was added 6 times of the sample volume to the sample tube. The tube was inverted 3 times and incubated at −20◦C in a freezer overnight. The next day, the sample tube was spun at 6000x g for 10 minutes. Then acetone was decanted, and the pellet was dried in a speed vacuum. Pellet was resuspended in ammonium bicarbonate 50mM, pH 8.0. Then 100 µg of total protein samples were re-suspended in 100 µl of ammonium bicarbonate (NH4HCO3). 100 µl of 0.05% RapigestTMSF was added to each sample. Samples were shaken using Vortex. Then the samples were concentrated to a volume of 100 µl using Vivaspin column MWCO 3000. After that, samples were centrifuged at 14000 rpm (20800 x g), for 10 – 15 minutes. The sample was then heated on a thermomixer at 80°C for 15 minutes. 5 µl of 100 mM DTT was added to each mixture and incubated at 37°C for 30 minutes in thermomixer. Then samples were added with 5 µl of 200 mM Iodoacetamide and incubated at room temperature for 45 minutes. 5 µl (0.2 µg/µl) of trypsin was then added to the reaction and each mixture was incubated overnight at 37°C. The trypsin digestion reaction was stopped by adding 1µl of concentrated trifluoroacetic acid (TFA) and incubated at 37°C for 20 minutes. The mixtures were then centrifuged at 14000 rpm (20800 x g), for 10 minutes. Supernatants were collected and stored at -80°C prior to use.
LCMS analysis
Peptide samples were evaporated down to 10 µl per sample. Each sample was mixed with 200 µl of formic acid and filtered using 0.45um regenerated cellulose membrane syringe filter. The LC-MS analysis was conducted using Orbitrap Fusion mass spectrometer coupled with Dionex 3000 Ultimate RSLCnano (Thermo Fisher Scientific) liquid chromatography system. EASY-Spray Column Acclaim PepMapTM C18 (100 A0, 2 µm particle size, 50 µm id x 15 cm) was used as the analytical column whereas Easy column C18 (2 cm, 0.1 mm i.d., 5 µm) was used as the pre-column. The MS2 spectra were analysed by ion trap MS (ITMS) using the following parameters: rapid scan rate with a resolving power of 60000, AGC target of 1.0e2 (100), 1.6 m/z isolation window, and a maximum injection time of 250 ms. Precursors were fragmented by collision-induced dissociation (CID) and high-energy collision dissociation (HCD) at normalised collision energy of 30% and 28%. Each sample was analysed thrice. Raw data obtained was analysed using Thermo ScientificTM Proteome DiscovererTM.
Bioinformatic analysis
Thermo ScientificTM Proteome DiscovererTM was used to analyse the peptide identified from raw data. There are another three types of bioinformatics tools employed to further analyse the data obtained. These include Perseus, Panther and Reactome. Based on the list of protein and its expression obtained from an analysis by Perseus, another analysis was done using Panther (Protein Analysis Through Evolutionary Relationships) software Version 13.0 (http://pantherb.org/). This is a classification system with a large curated biological database of biomolecules (genes, proteins or transcripts) families and their functionally related subfamilies. This tool is part of the Gene Ontology Reference Genome Project and is effective for high-throughput analysis. An analysis was done using this tool to explain the biological function, cellular localisation, molecular function, and protein class of identified protein obtained previously. Other than Perseus and Panther, another tool known as Reactome version 64 (http://reactome.org) was used in the analysis. Reactome is a curated pathway database that provides comprehensive analysis and interpretation of pathway knowledge. Significant pathways were chosen from the analysis according to the number of proteins involved in it and the nature of the pathways.
Results
Protein identification
After the raw mass spectrometry data were loaded into Thermo Scientific™ Proteome Discoverer™ Software Version 2.1, de novo sequencing and database search was performed by accessing the Uniprot_homo_sapiens, a public sequence database for protein identification purpose. There were 4392 peptide-spectrum matches (PSM), 539 peptide group, and 122 protein groups identified in the untreated samples. On the other hand, there were 3266 PSM, 412 peptide group and 102 protein groups identified in the S.asper-treated samples.
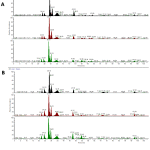 |
Figure 1: Total ion chromatograms (TICs) plot for the (A) untreated cells and (B) treated cells |
The total 224 proteins that were found in both cells (untreated and treated) were further analysed using bioinformatics data interpretation tool that is available online, Panther. This was done to obtain the information on proteins classification based on its biological process, molecular function, and cellular localisation. As shown in Figure 2.A, most of the proteins identified from untreated cells are involved in the cellular process (32.9%), followed by the metabolic process (25.3%), and cellular component organization or biogenesis (12.0%). While most of the proteins identified from treated cells are involved in the cellular process (32.6%). This is followed by a metabolic process (27.9%), and cellular component organization or biogenesis (12.4%). In Figure 2.B, most of the proteins identified from the untreated group are involved in binding (57%), followed by catalytic activity (18.6%), structural molecule activity (15.1%), translation regulator activity (3.5%), transporter activity (2.3%), antioxidant activity (1.2%), signal transducer activity (1.2%), and receptor activity (1.2%). On the other hand, for treated cells, most of the proteins that are involved in binding (56.6%). This is followed by catalytic activity (21.1%), structural molecule activity (13.2%), translation regulator activity (6.6%), transporter activity (1.3%), and antioxidant activity (1.3%). The cellular localisation for untreated cells is classified as shown in Figure 2.C. These include cell part (40.6%), organelle (32.3%), macromolecular complex (16.7%), membrane (6.3%) and extracellular region (3.1%), and synapse (1.0%). As for treated, most of the protein is involved in the cell part (43.3%). This is followed by organelle (32.8%), macromolecular complex (17.9%). Few of the protein is involved in the membrane (3.0%) and extracellular region (3.0%).
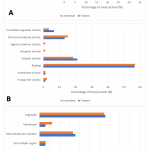 |
Figure 2: GO analysis illustrates classes of proteins differing between untreatedand treated cells. Protein with significant differences between the two groupswas subjected to GO classification in terms of (A) biological process, (B) molecular function, and (C) cellular localisation. |
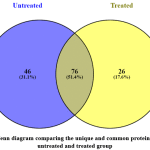 |
Figure 3: A Venn diagram comparing the unique and common proteins found in the untreated and treated group |
Protein quantification
Of 122 proteins identified in untreated groups, and 102 proteins identified in S.asper-treated group, only 76 proteins are common to both groups (Figure 3). These identified proteins were further analysed using a software namely Perseus. This software was used to identify the differentially expressed proteins in both groups. For this purpose, quantitative variations in protein abundance per injection between the protein group of the untreated and treated samples were represented by log2 ratios of normalised volume obtained by the Thermo Scientific™ Proteome Discoverer™ Software Version 2.1. The values were subjected to T-test (P<0.05).
Table 1: Differentially expressed proteins with at least 2-fold changes
| No | Protein Description | MW
[kDa] |
Fold Change | Regulation |
| 1. | Histone H4 | 11.36 | 3.03202 | Up |
| 2. | Plasminogen activator inhibitor 1 RNA-binding protein | 44.938 | 2.60309 | Up |
| 3. | Galectin-1 | 14.706 | 2.19671 | Up |
| 4. | 10 kDa heat shock protein, mitochondrial | 10.925 | 2.16321 | Up |
| 5. | Isoform 2 of Dermcidin | 12.406 | 2.01046 | Down |
| 6. | Keratinocyte proline-rich protein | 64.093 | 2.02556 | Down |
| 7. | Nascent polypeptide-associated complex subunit alpha, muscle-specific form | 205.295 | 2.03476 | Down |
| 8. | Serum albumin | 69.321 | 2.11567 | Down |
| 9. | Keratin, type I cytoskeletal 9 | 62.027 | 2.14074 | Down |
| 10. | Tropomyosin alpha-4 chain | 28.504 | 2.15584 | Down |
| 11. | Myristoylated alanine-rich C-kinase | 31.536 | 2.83294 | Down |
| 12. | Isoform 3 of Tumour protein D52 | 26.367 | 2.86387 | Down |
| 13. | Folate receptor alpha | 29.799 | 2.89709 | Down |
| 14. | Parathymosin | 11.523 | 3.58596 | Down |
Upon analysis, 54 of them were found differentially expressed. Of that, 14 proteins were up-regulated, 5 proteins appeared with no changes, and 35 proteins were down-regulated. The list of 14 differentially expressed proteins with more than 2-fold changes was listed as in Table 1.
Pathways analysis
Pathway enrichment analysis was carried out on common proteins with at least 2 peptides. The analysis was done using Reactome, an online bioinformatics tool. From that, 9 pathways that play a meaningful role in cancer were selected as listed in Table 2.
Table 2: Pathways to which the common proteins found were annotated.
| No | Pathway Name | Number of Proteins | P-Value |
| 1. | Metabolism of proteins | 13 | 0.167477704 |
| 2. | Post-translational protein modification | 11 | 0.07638734 |
| 3. | Signalling by Rho GTPases | 7 | 0.00443948 |
| 4. | Signalling by NOTCH | 5 | 0.003193056 |
| 5. | Cell Cycle | 5 | 0.183772998 |
| 6. | Cellular Senescence | 4 | 0.008353885 |
| 7. | Signalling by WNT | 4 | 0.001169484 |
| 8. | Transcriptional Regulation by TP53 | 4 | 0.102837272 |
| 9. | Cellular responses to stress | 4 | 0.136380287 |
Discussion
The MS analysis revealed 76 common proteins found in both groups. Of this, several proteins were selected according to its relationship with cancers as discussed in the past literature. S.asper mediated the upregulation of galectin-1 and 10 kDa heat shock protein. It is also found to downregulate the expression of dermcidin, keratin, type I, tropomyosin alpha-4 chain, myristoylated alanine-rich C-kinase, tumour protein D52, folate receptor alpha, and parathymosin.
Galectins, in general, are β-galactoside specific endogenous lectins with low molecular weight. It plays a role in cell growth, cell activation, and cell-cell, cell-matrix adhesion which include binding to carcinoembryonic antigen, laminin and metalloproteinase.39 Prototype galectin which has single carbohydrate recognition domain (CRD), comes in different forms and these include galectin-1, galectin-2, galectin-5, galectin-7, galectin-10, galectin-11, galectin-13, galectin-14, galectin -15.40 Galectin-1 is found overexpressed in many forms of human tumour. Louka, et al., (2017) discovered that galectin-1 is upregulated in breast cancer compared to benign breast lesion.41 The study observed the elevation of galectin-1 expression corresponds to the increased activity of MMP-2 and MMP-9. Matrix metalloproteinases (MMP) is typically responsible for the degradation of most extracellular matrix proteins during organogenesis, growth and normal tissue turnover.42 This means that the increased activity of galectin-1 may serve as a way for cancer metastasis and invasion. Galectin-1 was also found increased in colon cancer, liver cancer, pancreatic cancer, and cervical cancer.43,44,45,46 In cervical cancer, overexpression of galectin-1 often associated with invasion and metastasis.47 In our recent study, galectin-1 was found increased in abundance after treatment with S.asper. This indicated that galectin-1 was initially low in cervical cancer which seems consistent with studies that found galectin-1 to be under-expressed in head and neck squamous cell cancer and uterine cancer.48,49
Heat shock proteins (HSPs) are chaperone that play significant role in protecting its client protein from being degraded, hypoxic, thermally and oxidatively stressed.50 HSPs are categorized into six families depending on their relative molecular sizes. These include HSP27, HSP40, HSP60, HSP70, HSP90, and family of large HSPs (HSP110 and HSP170).51,52,53 The chaperonage function is mediated by facilitating correction of the misfolded proteins, maintaining the innate structure and function of their client protein.54,55 In the case of cancer, HSPs are considered as the regulators as they protect oncoprotein associated with cancer proliferation, differentiation, and progression. HSP10 which was identified in this study is a chaperone located in the mitochondria. Past literature reported that overexpression of HSP10 is found in several tumours like exocervical carcinoma, large bowel and uterine exocervix, and serous ovarian carcinoma. The studies discovered that elevated HSP10 level is associated with carcinogenesis and this chaperone protein employ different mechanisms to influence tumour initiation and progression.56,57,58 Contrary to expectation, we found HSP10 to be initially low in HeLa cells compared to its level after S.asper treatment. In other words, S.asper seems to cause HSP10 to be up-regulated in HeLa cells. The detailed mechanism that results in this pattern of expression remains elusive.
Dermcidin has been found to act as a growth and survival factor in breast cancer.59 This is concluded when overexpression of dermcidin increases cell proliferation and cell resistance to oxidative stress. This finding substantiates Brauer et al. (2014)’s work, where a significantly high level of dermicidin was found corresponded with an early progression of N-methylnitrosourea-induced breast cancer.60 In a different study, the oncogenic effect of dermcidin in breast cancer was found exerted via ERBB signalling.61 The result demonstrated a reduction of dermcidin level upon S.asper treatment. S.asper seems to cause suppression of dermcidin oncogenic effects which include cancer growth and progression. However, the evidence regarding the expression of dermcidin in cervical cancer is still limited.
Based on past reports, there are 12 keratins identified in cervical carcinoma. These include keratin 4, 5, 6, 7, 8, 10, 13, 14, 16, 17, 18, and 19.62 Expression of keratin was also found in breast cancer. According to the study, ninety per cent of all their breast cancer samples have had the expression of keratin 7, 8, 18, and 19.63 Keratins have been found contributing to cell size determination, translation control, proliferation, cell type-specific, organelle transport, malignant transformation and various stress responses.64 In cancer cells, studies confirm that keratins actively involved in cancer cell progression, metastasis, and response to treatments.65,66 This support a finding of which overexpression of keratin particularly keratin-18 presence in breast cancer correlates with poor prognosis of the disease.67 Oppositely, down-regulation of keratins provide a sign of improvement and better prognosis for cancer. This pattern of expression can be seen in our study whereby S.asper exhibit the ability to suppress the expression of keratins in cervical cancer.
Another protein known as tropomyosin alpha-4 chain was differentially expressed in our study. Tropomyosin plays a crucial role in regulating the contraction of the muscle.68,69 The expression of tropomyosin has been considered as varies depending on its subtype and the type of tumour. Pieces of evidence explain that tropomyosin-4 is up-regulated in oesophageal carcinoma and colon cancer.70,71 In a different study, Pawlak and co-investigators (2004) found that the level of tropomyosin-1 and -2 were markedly reduced in urinary bladder carcinoma.72 Bae et al. in 2005 identified the presence of tropomyosin in squamous cervical cancer but no difference in expression pattern was observed between cancerous tissue and normal cervical tissue.73 In a more recent study, tropomyosin-4 was proved lowly expressed in invasive squamous cervical carcinoma compared to Cervical Intraepithelial Neoplasia (CIN) and normal epithelium.74 Hence more molecular explanation is required to acquire better insight on the association of tropomyosin-4 and cervical cancer.
Myristoylated Alanine-Rich C-Kinase Substrate (MARCKS) protein has been found involved crucially in cell development and regeneration.75 Upregulation of MARCKS protein increases the activity of cell motility. Besides, it was found associated with the development of intimal hyperplasia in the murine carotid ligation model.76 The increased activity of MARCKS has been seen to positively correlated with the development of lung cancer in advanced stages (stage II-IV), lymph node metastatic status, and malignant phenotypes.77 In hepatocellular carcinoma cell line (HepG2), MARCKS protein was observed to have a close relationship with cell migration and invasion. This was concluded when knockdown of MARCKS significantly reduced HepG2 migration and invasion activities.78 MARCKS protein is a key regulator in the expression of micro-RNA 21 (miR-21) which found to enhance apoptosis resistance, motility, and invasion in prostate cancer cells.79 A study found that downregulation of MARCKS resulted in a possible anti-tumour effect at the metastatic site of colon carcinoma.80 S.asper treatment was found able to suppress the expression of MARCKS protein. This might potentially contribute to the reduction of metastasis and invasion activity of cervical cancer.
Another protein found to be altered by S.asper is tumour protein D52 (TPD52). TPD52 is a protein that is overexpressed in ovarian cancer.81 The expression is ranging from absent in benign tumour to overexpression in all invasive samples. TPD52 was also found overexpressed in prostate cancer. The knockdown of TPD52 was reported to lead to an inducement of apoptosis through a caspase-dependent pathway. Furthermore, transient overexpression of EGFP-TPD52 results in the increased proliferation rate of LNCap cells.82 In a different report, TPD52 has been witnessed as being overexpressed in all cases of colorectal cancer with significantly more than 3-fold change.83 Besides, a similar pattern of expression was also seen in breast cancer.84 The findings reported by previous literature are predominantly consistent with ours in which TPD52 expression is high in cancer cells. Upon treatment with S.asper, the expression was observed to decrease up to more than 2-fold change. However, the details about the target function associated with TPD52 overexpression remain unclear.
Folate receptor alpha (FRα) is a protein that is bound at the glycosylphosphatidylinositol (GPI) anchor of the cell membrane.85 It provides a high-affinity binding site for folate. Studies show that in the condition of folate deficiency, FRα tends to over-expressed.86 In accordance with that, constant folate deficiency increases the risk for cervical carcinogenesis.87 FRα has been proved to be overexpressed in epithelial ovarian cancer.88 This is supported by a more recent study which produces a similar result in metastatic foci and recurrent ovarian cancer.89 FRα was also found positively correlated in adenocarcinoma of non-small lung cancer.90 This finding accords with another study conducted by Nunez et al. published in the same year.91 Not just that, FRα was also demonstrated to be upregulated in 30% of breast cancer cases and 70-80% of stage VI triple-negative breast cancer cases suggesting it to be a promising therapeutic target.90 In addition, overexpression of FRα is proved to be associated with poor outcome in breast cancer.92 The reduction of FRα expression in response to treatment in this study suggested that S.asper may potentially implicate certain poorly understood mechanism related to the anti-folate receptor and cervical malignancy.
It was analysed that the positive expression of parathymosin may be associated with poor prognosis of squamous cell carcinoma and adenocarcinoma of the gallbladder.93 A study found that parathymosin was involved in promoting cell proliferation by regulating the level of glucocorticoids.94 In gastric epithelium, parathymosin expression was seen to be altered in tumour compared to normal gastric.95 Parathymosin level was also observed to be increased in human upper urinary tract transitional cell carcinoma.96 In a more recent report, parathymosin was suggested to be a potential predictor for early recurrence and poor prognosis of hepatocellular carcinoma taken into consideration of its overexpression pattern in 71% of the cases.97 Our finding, on the other hand, demonstrated the ability of S.asper to suppress parathymosin activity up to 3-fold change. This suggests that parathymosin may become one of the key therapeutic targets in S.asper treatment mechanism of cervical cancer.
Conclusion
In the present study, we have presented several proteins that are identified and quantified from the S.asper-treated and untreated samples of HeLa cervical cancer cells. Nine proteins were selected as they have been discussed in many past literatures to be associated directly or indirectly to cancers. Based on previous studies, seven of the proteins seem consistent to support our hypothesis while only two seem to be contradictory. Taken together, these findings would likely suggesting that S.asper demonstrated significant anticancer effects on cervical cancer cell line through the regulation of several key proteins and pathways. Hence, an additional investigation needs to be conducted to confirm the involvement of each protein in the S.asper treatment on cervical cancer. Besides, further study is also required to elucidate comprehensively the cellular and molecular mechanism which contribute to the treatment effects.
Conflict of interest
The authors declare that there are no conflicts of interest.
Acknowledgement
The authors would like to thank the Malaysian Ministry of Education and Universiti Teknologi MARA for funding the study under National Research Acculturation Grant Scheme (RAGS) (Ref no.: RAGS/1/2015/SKK08/UITM/03/1). The authors gratefully thank Advanced Medical and Dental Institute (AMDI), Universiti Sains Malaysia (USM) and Universiti Teknologi MARA (UiTM) Penang Branch, for supportively providing spaces and sufficient facilities throughout the study process.
References
- Bruni, L, Albero G, Serrano B, Mena M, Gómez D, Muñoz J, and others, ‘Human Papillomavirus and Related Diseases in Malaysia’, ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre), 2018
- Torre, Lindsey A, Rebecca L Siegel, Elizabeth M Ward, and Ahmedin Jemal, ‘Global Cancer Incidence and Mortality Rates and Trends–An Update.’, Cancer Epidemiology, Biomarkers & Prevention : A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology, 2016; 25: 16–27
- Schiffman, Mark, Philip E Castle, Jose Jeronimo, Ana C Rodriguez, and Sholom Wacholder, ‘Human Papillomavirus and Cervical Cancer.’, Lancet (London, England), 2007; 370: 890–907
- Wright, Jason D., Jianduan Li, Daniela S. Gerhard, Zhengyan Zhang, Phyllis C. Huettner, Matthew A. Powell, and others, ‘Human Papillomavirus Type and Tobacco Use as Predictors of Survival in Early Stage Cervical Carcinoma’, Gynecologic Oncology, 2005; 98: 84–91
- Moreno, Victor, F Xavier Bosch, Nubia Muñoz, Chris J L M Meijer, Keerti V Shah, Jan M M Walboomers, and others, ‘Effect of Oral Contraceptives on Risk of Cervical Cancer in Women with Human Papillomavirus Infection : The IARC Multicentric Case- Control Study’, 2002; 359: 1085–1092
- Xu, H., S. Egger, L. S Velentzis, D. L O’Connell, E. Banks, J. Darlington-Brown, and others, ‘Hormonal Contraceptive Use and Smoking as Risk Factors for High-Grade Cervical Intraepithelial Neoplasia in Unvaccinated Women Aged 30–44 Years: A Case-Control Study in New South Wales, Australia’, Cancer Epidemiology, 2018; 55: 162–169
- Appleby, P, V Beral, A Berrington de Gonzalez, D Colin, S Franceschi, A Goodhill, and others, ‘Cervical Cancer and Hormonal Contraceptives: Collaborative Reanalysis of Individual Data for 16,573 Women with Cervical Cancer and 35,509 Women without Cervical Cancer from 24 Epidemiological Studies’, Lancet, 2007; 370: 1609–1621
- Rizzo, Anthony E., and Sarah Feldman, ‘Update on Primary HPV Screening for Cervical Cancer Prevention’, Current Problems in Cancer, 2018; 0: 1–14
- zur Hausen, Harald, ‘Papillomaviruses and Cancer: From Basic Studies to Clinical Application’, Nature Reviews Cancer, 2002; 2: 342
- Ramakrishnan, Sabitha, Steena Partricia, and Ganeshan Mathan, ‘ScienceDirect Overview of High-Risk HPV ’ s 16 and 18 Infected Cervical Cancer : Pathogenesis to Prevention’, Biomedicine et Pharmacotherapy, 2015; 70: 103–110
- Sankaranarayanan, Rengaswamy, Smita Joshi, Richard Muwonge, Pulikottil Okkuru Esmy, Partha Basu, Priya Prabhu, and others, ‘Can a Single Dose of Human Papillomavirus (HPV) Vaccine Prevent Cervical Cancer? Early Findings from an Indian Study.’, Vaccine, 2018; 36: 4783–4791
- Levin, Ann, Susan A. Wang, Carol Levin, Vivien Tsu, and Raymond Hutubessy, ‘Costs of Introducing and Delivering HPV Vaccines in Low and Lower Middle Income Countries: Inputs for GAVI Policy on Introduction Grant Support to Countries’, PLoS ONE, 2014; 9
- Laurent, Jessica St., Rebecca Luckett, and Sarah Feldman, ‘HPV Vaccination and the Effects on Rates of HPV-Related Cancers’, Current Problems in Cancer, 2018; 42: 493–506
- Nickel, Brooke, Rachael H Dodd, Robin M Turner, Jo Waller, Laura Marlow, Gregory Zimet, and others, ‘Factors Associated with the Human Papillomavirus ( HPV ) Vaccination across Three Countries Following Vaccination Introduction’, 2017; 8: 169–176
- Bonanni, Paolo, Beatrice Zanella, Francesca Santomauro, Chiara Lorini, Angela Bechini, and Sara Boccalini, ‘Safety and Perception: What Are the Greatest Enemies of HPV Vaccination Programmes?’, Vaccine, 2018; 36: 5424–5429
- World Health Organization, WHO Guidelines for Screening and Treatment of Precancerous Lesions for Cervical Cancer Prevention (World Health Organization, 2013)
- Raffle, A E, B Alden, M Quinn, P J Babb, and M T Brett, ‘Outcomes of Screening to Prevent Cancer: Analysis of Cumulative Incidence of Cervical Abnormality and Modelling of Cases and Deaths Prevented.’, BMJ (Clinical Research Ed.), 2003; 326: 901
- Vicus, Danielle, Rinku Sutradhar, Yan Lu, Laurie Elit, Rachel Kupets, and Lawrence Paszat, ‘The Association between Cervical Cancer Screening and Mortality from Cervical Cancer: A Population Based Case-Control Study.’, Gynecologic Oncology, 2014; 133: 167–171
- Yoshida, Y, C L Schmaltz, J Jackson-Thompson, and E J Simoes, ‘The Impact of Screening on Cancer Incidence and Mortality in Missouri, USA, 2004-2013.’, Public Health, 2018; 154: 51–58
- Priaulx, Jennifer, Harry J de Koning, Inge M C M de Kok, György Széles, and Martin McKee, ‘Identifying the Barriers to Effective Breast, Cervical and Colorectal Cancer Screening in Thirty One European Countries Using the Barriers to Effective Screening Tool (BEST)’, Health Policy, 2018
- Lobo, Niyati, Meghana Kulkarni, Simon Hughes, Rajesh Nair, Muhammad Shamim Khan, and Ramesh Thurairaja, ‘Urologic Complications Following Pelvic Radiotherapy.’, Urology, 2018; 122: 1–9
- Evans, Elin, and John Staffurth, ‘Principles of Cancer Treatment by Radiotherapy’, Surgery (United Kingdom), 2018; 36: 134–38
- Overbeek, Annelies, Marleen H. van den Berg, Flora E. van Leeuwen, Gertjan J.L. Kaspers, Cornelis B. Lambalk, and Eline van Dulmen-den Broeder, ‘Chemotherapy-Related Late Adverse Effects on Ovarian Function in Female Survivors of Childhood and Young Adult Cancer: A Systematic Review’, Cancer Treatment Reviews, 2017; 53: 10–24
- Gunderson, Camille C., Ursula Matulonis, and Kathleen N. Moore, ‘Management of the Toxicities of Common Targeted Therapeutics for Gynecologic Cancers’, Gynecologic Oncology, 2018; 148: 591–600
- Rastogi, Subha, Dinesh K. Kulshreshtha, and Ajay Kumar Singh Rawat, ‘Streblus Asper Lour. (Shakhotaka): A Review of Its Chemical, Pharmacological and Ethnomedicinal Properties’, Evidence-Based Complementary and Alternative Medicine, 2006; 3: 217–222
- Seeni, Azman, Nur Ayunie, and Ridhwan Abdul Wahab, ‘Apoptosis Inducer from Streblus Asper Extracts for Cancer Chemoprevention’, in Novel Apoptotic Regulators in Carcinogenesis, 2012; 1–25
- Chatterjee, Ranjit Kumar, Nigar Fatma, Puvvada Kalpana Murthy, Preeti Sinha, Dinesh Kumar Kulshrestha, and Bhola Nath Dhawan, ‘Macrofilaricidal Activity of the Stembark of Streblus Asper and Its Major Active Constituents’, Drug Development Research, 1992; 26: 67–78
- Taweechaisupapong, S., T. Choopan, S. Singhara, S. Chatrchaiwiwatana, and S. Wongkham, ‘In Vitro Inhibitory Effect of Streblus Asper Leaf-Extract on Adhesion of Candida Albicans to Human Buccal Epithelial Cells’, Journal of Ethnopharmacology, 2005; 96: 221–226
- Taweechaisupapong, S, P Klanrit, S Singhara, W Pitiphat, and S Wongkham, ‘Inhibitory Effect of Streblus Asper Leaf-Extract on Adhesion of Candida Albicans to Denture Acrylic.’, Journal of Ethnopharmacology, 2006; 106: 414–417
- Sripanidkulchai, Bungorn, Jintana Junlatat, Nawarat Wara-aswapati, and Doosadee Hormdee, ‘Anti-Inflammatory Effect of Streblus Asper Leaf Extract in Rats and Its Modulation on Inflammation-Associated Genes Expression in RAW 264 . 7 Macrophage Cells’, 2009; 12: 566–570
- Taweechaisupapong, Suwimol, Sopit Wongkham, and Supaporn Chareonsuk, ‘Selective Activity of Streblus Asper on Mutans Streptococci’, 2000; 70: 73–79
- Chen, Hong, Jun Li, Qiang Wu, Xiao Tao Niu, Mao Tong Tang, Xin Lan Guan, and others, ‘Anti-HBV Activities of Streblus Asper and Constituents of Its Roots’, Fitoterapia, 2012; 83: 643–649
- Li, Jun, Yan Huang, Xin-lan Guan, Jian Li, Sheng-ping Deng, and Qiang Wu, ‘Phytochemistry Anti-Hepatitis B Virus Constituents from the Stem Bark of Streblus Asper’, 2012; 82: 100–109
- Li, Jun, Ai-Ping Meng, Xin-Lan Guan, Jian Li, Qiang Wu, Sheng-Ping Deng, and others, ‘Anti-Hepatitis B Virus Lignans from the Root of Streblus Asper.’, Bioorganic & Medicinal Chemistry Letters, 2013; 23: 2238–2244
- Li, Lu-Qing, Jun Li, Yan Huang, Qiang Wu, Sheng-Ping Deng, Xiao-Jian Su, and others, ‘Lignans from the Heartwood of Streblus Asper and Their Inhibiting Activities to Hepatitis B Virus’, Fitoterapia, 2012; 83: 303—309
- Kumar, R. B.Suresh, Biswakanth Kar, Narayan Dolai, Asis Bala, and Pallab Kanti Haldar, ‘Evaluation of Antihyperglycemic and Antioxidant Properties of Streblus Asper Lour against Streptozotocin-Induced Diabetes in Rats’, Asian Pacific Journal of Tropical Disease, 2012; 2:139–143
- Choudhury, Monjoy Kumar, S. Venkatraman, and Lokesh Upadhyay, ‘Phytochemical Analysis and Peripheral Glucose Utilization Activity Determination of Steblus Asper’, Asian Pacific Journal of Tropical Biomedicine, 2012; 2: 1–3
- Phutdhawong, Weerachai, Arworn Donchai, John Korth, Stephen G Pyne, Porntipa Picha, Jarunya Ngamkham, and others, ‘The Components and Anticancer Activity of the Volatile Oil from Streblus Asper’, Flavour and Fragrance Journal, 2004; 19: 445–447
- Sujathan, K, Thara Somanathan, and Prathapan Remani, ‘Expression of Galectin-1, Galectin-3 and T-Antigen in Breast Carcinoma Tissues and Its Significance in Axillary Lymph Node Infiltration’, Recent Research in Science and Technology, 2011; 3: 85–90
- Wang, Lufang, Yanyan Zhao, Yanshi Wang, and Xin Wu, ‘The Role of Galectins in Cervical Cancer Biology and Progression’, 2018
- Louka, Manal, Hebatallah Said, Sara El Sayed, and Mohamed El-Shinawi, ‘Galectin 1 Overexpression in Breast Cancer Tissues: Relation to Serum Matrix Metalloproteinase 2 and 9 Activity’, Gene Reports, 2017; 7: 184–187
- Sorsa, T, L Tjaderhane, and T Salo, ‘Matrix Metalloproteinases (MMPs) in Oral Diseases.’, Oral Diseases, 2004; 10: 311–318
- Sanjuan, X, P L Fernandez, A Castells, V Castronovo, F van den Brule, F T Liu, and others, ‘Differential Expression of Galectin 3 and Galectin 1 in Colorectal Cancer Progression.’, Gastroenterology, 1997; 113: 1906–15
- Spano, Daniela, Roberta Russo, Vittorio Di Maso, Natalia Rosso, Luigi M Terracciano, Massimo Roncalli, and others, ‘Galectin-1 and Its Involvement in Hepatocellular Carcinoma Aggressiveness.’, Molecular Medicine (Cambridge, Mass.), 2010; 16: 102–115
- Chen, R, S Pan, N A Ottenhof, R F De Wilde, C L Wolfgang, Z Lane, and others, ‘Stromal Galectin-1 Expression Is Associated with Long-Term Survival in Resectable Pancreatic Ductal Adenocarcinoma’, Cancer Biology and Therapy, 2012; 13: 899–907
- Kohrenhagen, N, H U Volker, M Kapp, J Dietl, and U Kammerer, ‘Increased Expression of Galectin-1 during the Progression of Cervical Neoplasia.’, International Journal of Gynecological Cancer : Official Journal of the International Gynecological Cancer Society, 2006; 16: 2018–2022
- Punt, Simone, Victor L Thijssen, Johannes Vrolijk, Cornelis D de Kroon, Arko Gorter, and Ekaterina S Jordanova, ‘Galectin-1, -3 and -9 Expression and Clinical Significance in Squamous Cervical Cancer’, PLOS ONE, 2015; 10: 1–13
- Choufani, G, N Nagy, S Saussez, H Marchant, P Bisschop, M Burchert, and others, ‘The Levels of Expression of Galectin-1, Galectin-3, and the Thomsen- Friedenreich Antigen and Their Binding Sites Decrease as Clinical Aggressiveness Increases in Head and Neck Cancers’, Cancer, 1999; 86: 2353–2363
- Makino, Kenichi, Kazuhiro Kawamura, Wataru Sato, Nanami Kawamura, Toshio Fujimoto, and Yukihiro Terada, ‘Inhibition of Uterine Sarcoma Cell Growth through Suppression of Endogenous Tyrosine Kinase B Signaling.’, PloS One, 2012; 7: 1-9
- Jia, Haibo, Amadou I Halilou, Liang Hu, Wenqian Cai, Jing Liu, and Bo Huang, ‘Heat Shock Protein 10 (Hsp10) in Immune-Related Diseases: One Coin, Two Sides.’, International Journal of Biochemistry and Molecular Biology, 2011; 2: 47–57
- Ciocca, Daniel R, and Stuart K Calderwood, ‘Heat Shock Proteins in Cancer: Diagnostic, Prognostic, Predictive, and Treatment Implications.’, Cell Stress & Chaperones, 2005; 10: 86–103
- Wang, J.-T., L Ding, S.-W. Jiang, J Hao, W.-M. Zhao, Q Zhou, and others, ‘Folate Deficiency and Aberrant Expression of DNA Methyltransferase 1 Were Associated with Cervical Cancerization’, Current Pharmaceutical Design, 2014; 20: 1639–1646
- Wu, Jianming, Tuoen Liu, Zechary Rios, Qibing Mei, Xiukun Lin, and Shousong Cao, ‘Heat Shock Proteins and Cancer’, Trends in Pharmacological Sciences, 2017; 38: 226–256
- Liu, Tuoen, Christopher K Daniels, and Shousong Cao, ‘Comprehensive Review on the HSC70 Functions, Interactions with Related Molecules and Involvement in Clinical Diseases and Therapeutic Potential.’, Pharmacology & Therapeutics, 2012; 136: 354–374
- Macario, Alberto J L, and Everly Conway de Macario, ‘Molecular Chaperones: Multiple Functions, Pathologies, and Potential Applications.’, Frontiers in Bioscience : A Journal and Virtual Library, 2007; 12: 2588–2600
- Cappello, Francesco, Marianna Bellafiore, Sabrina David, Rita Anzalone, and Giovanni Zummo, ‘Ten Kilodalton Heat Shock Protein (HSP10) Is Overexpressed during Carcinogenesis of Large Bowel and Uterine Exocervix.’, Cancer Letters, 196 (2003; 196: 35–41
- Cappello, Francesco, ‘HSP60 and HSP10 as Diagnostic and Prognostic Tools in the Management of Exocervical Carcinoma.’, Gynecologic Oncology 2003; 661
- Tetu, Bernard, Ion Popa, Isabelle Bairati, Sylvain L’Esperance, Magdalena Bachvarova, Marie Plante, and others, ‘Immunohistochemical Analysis of Possible Chemoresistance Markers Identified by Micro-Arrays on Serous Ovarian Carcinomas.’, Modern Pathology : An Official Journal of the United States and Canadian Academy of Pathology, Inc, 2008; 21: 1002–1010
- Porter, Dale, Stanislawa Weremowicz, Koei Chin, Pankaj Seth, Aparna Keshaviah, Jaana Lahti-Domenici, and others, ‘A Neural Survival Factor Is a Candidate Oncogene in Breast Cancer.’, Proceedings of the National Academy of Sciences of the United States of America, 2003;100: 10931–10936
- Brauer, Heather Ann, Monica D’Arcy, Tanya E Libby, Henry J Thompson, Yutaka Y Yasui, Nobuyuki Hamajima, and others, ‘Dermcidin Expression Is Associated with Disease Progression and Survival among Breast Cancer Patients.’, Breast Cancer Research and Treatment, 2014;144: 299–306
- Bancovik, Jasna, Dayson F Moreira, Daniel Carrasco, Jun Yao, Dale Porter, Ricardo Moura, and others, ‘Dermcidin Exerts Its Oncogenic Effects in Breast Cancer via Modulation of ERBB Signaling.’, BMC Cancer, 2015; 15: 70
- Smedts, F, F Ramaekers, S Troyanovsky, M Pruszczynski, M Link, B Lane, and others, ‘Keratin Expression in Cervical Cancer.’, The American Journal of Pathology, 1992; 141: 497–511
- Shao, Mu-Min, Siu Ki Chan, Alex M C Yu, Christopher C F Lam, Julia Y S Tsang, Philip C W Lui, and others, ‘Keratin Expression in Breast Cancers.’, Virchows Archiv : An International Journal of Pathology, 2012; 461: 313–322
- Magin, Thomas M, Preethi Vijayaraj, and Rudolf E Leube, ‘Structural and Regulatory Functions of Keratins.’, Experimental Cell Research, 2007; 313: 2021–2032
- Alix-Panabières, Catherine, Jean-Pierre Vendrell, Monique Slijper, Olivier Pellé, Eric Barbotte, Grégoire Mercier, and others, ‘Full-Length Cytokeratin-19 Is Released by Human Tumor Cells: A Potential Role in Metastatic Progression of Breast Cancer’, Breast Cancer Research, 11 2009; R39
- Ding, Shi-Jian, Yan Li, Ye-Xiong Tan, Man-Rong Jiang, Bo Tian, Ying-Kun Liu, and others, ‘From Proteomic Analysis to Clinical Significance’, Molecular & Cellular Proteomics, 2004; 3: 73–81
- Somiari, Richard I, Anthony Sullivan, Stephen Russell, Stella Somiari, Hai Hu, Rick Jordan, and others, ‘High-Throughput Proteomic Analysis of Human Infiltrating Ductal Carcinoma of the Breast.’, Proteomics, 2003;3: 1863–1873
- Geeves, Michael A, Sarah E Hitchcock-DeGregori, and Peter W Gunning, ‘A Systematic Nomenclature for Mammalian Tropomyosin Isoforms.’, Journal of Muscle Research and Cell Motility, 2015; 36: 147–153
- Gunning, Peter W, Edna C Hardeman, Pekka Lappalainen, and Daniel P Mulvihill, ‘Tropomyosin – Master Regulator of Actin Filament Function in the Cytoskeleton.’, Journal of Cell Science, 2015; 128: 2965–2674
- Harada, Toshio, Yasuhiro Kuramitsu, Akira Makino, Masanori Fujimoto, Norio Iizuka, Yoshinobu Hoshii, and others, ‘Expression of Tropomyosin Alpha 4 Chain Is Increased in Esophageal Squamous Cell Carcinoma as Evidenced by Proteomic Profiling by Two-Dimensional Electrophoresis and Liquid Chromatography-Mass Spectrometry/Mass Spectrometry.’, Proteomics. Clinical Applications, 2007; 1: 215–223
- Yang, Rui, Gang Zheng, Defa Ren, Chunzhou Chen, Cheng Zeng, Wei Lu, and others, ‘The Clinical Significance and Biological Function of Tropomyosin 4 in Colon Cancer’, Biomedicine & Pharmacotherapy, 2018; 101: 1–7
- Pawlak, Geraldine, Terence W McGarvey, Trang B Nguyen, John E Tomaszewski, Raghunath Puthiyaveettil, S Bruce Malkowicz, and others, ‘Alterations in Tropomyosin Isoform Expression in Human Transitional Cell Carcinoma of the Urinary Bladder.’, International Journal of Cancer, 2004;110: 368–373
- Bae, Su Mi, Chang-Hun Lee, Young Lae Cho, Kye Hyun Nam, Yong Wan Kim, Chong Kook Kim, and others, ‘Two-Dimensional Gel Analysis of Protein Expression Profile in Squamous Cervical Cancer Patients.’, Gynecologic Oncology, 2005; 99: 26–35
- Lomnytska, M I, S Becker, I Bodin, A Olsson, K Hellman, A-C Hellstrom, and others, ‘Differential Expression of ANXA6, HSP27, PRDX2, NCF2, and TPM4 during Uterine Cervix Carcinogenesis: Diagnostic and Prognostic Value.’, British Journal of Cancer, 2011;104: 110–119
- El Amri, Mohamed, Una Fitzgerald, and Gerhard Schlosser, ‘MARCKS and MARCKS-like Proteins in Development and Regeneration’, Journal of Biomedical Science, 2018; 25: 43
- Yu, Dan, George Makkar, Dudley K Strickland, Thomas A Blanpied, Deborah J Stumpo, Perry J Blackshear, and others, ‘Myristoylated Alanine-Rich Protein Kinase Substrate (MARCKS) Regulates Small GTPase Rac1 and Cdc42 Activity and Is a Critical Mediator of Vascular Smooth Muscle Cell Migration in Intimal Hyperplasia Formation.’, Journal of the American Heart Association, 2015; 4: e002255
- Chen, Ching-Hsien, Chun-Lung Chiu, Kenneth B Adler, and Reen Wu, ‘A Novel Predictor of Cancer Malignancy: Up-Regulation of Myristoylated Alanine-Rich C Kinase Substrate Phosphorylation in Lung Cancer.’, American Journal of Respiratory and Critical Care Medicine, 2014; 1002–1004
- Song, Jianjun, Qi Wang, Yongyun Luo, Peng Yuan, Chaofeng Tang, Yongfeng Hui, and others, ‘MiR-34c-3p Inhibits Cell Proliferation, Migration and Invasion of Hepatocellular Carcinoma by Targeting MARCKS.’, International Journal of Clinical and Experimental Pathology, 2015; 8: 12728–12737
- Li, Tao, Dong Li, Jianjun Sha, Peng Sun, and Yiran Huang, ‘MicroRNA-21 Directly Targets MARCKS and Promotes Apoptosis Resistance and Invasion in Prostate Cancer Cells.’, Biochemical and Biophysical Research Communications, 2009; 383: 280–285
- Rombouts, Krista, Vinicio Carloni, Tommaso Mello, Sara Omenetti, Sara Galastri, Stefania Madiai, and others, ‘Myristoylated Alanine-Rich Protein Kinase C Substrate (MARCKS) Expression Modulates the Metastatic Phenotype in Human and Murine Colon Carcinoma in Vitro and in Vivo.’, Cancer Letters, 2013; 333: 244–252
- Byrne, Jennifer A, Rosemary L Balleine, Marlena Schoenberg Fejzo, Janelle Mercieca, Yoke-Eng Chiew, Yael Livnat, and others, ‘Tumor Protein D52 (TPD52) Is Overexpressed and a Gene Amplification Target in Ovarian Cancer.’, International Journal of Cancer, 2005; 117: 1049–1054
- Ummanni, Ramesh, Steffen Teller, Heike Junker, Uwe Zimmermann, Simone Venz, Christian Scharf, and others, ‘Altered Expression of Tumor Protein D52 Regulates Apoptosis and Migration of Prostate Cancer Cells.’, The FEBS Journal, 2008;275: 5703–5713
- Petrova, Darinka Todorova, Abdul R Asif, Victor W Armstrong, Ivanka Dimova, Svetoslav Toshev, Nikolay Yaramov, and others, ‘Expression of Chloride Intracellular Channel Protein 1 (CLIC1) and Tumor Protein D52 (TPD52) as Potential Biomarkers for Colorectal Cancer.’, Clinical Biochemistry, 2008;41: 1224–1236
- Balleine, R L, M S Fejzo, P Sathasivam, P Basset, C L Clarke, and J A Byrne, ‘The HD52 (TPD52) Gene Is a Candidate Target Gene for Events Resulting in Increased 8q21 Copy Number in Human Breast Carcinoma’, Genes Chromosomes and Cancer, 2000;29: 48–57
- Zhao, Rongbao, and I David Goldman, ‘Folate and Thiamine Transporters Mediated by Facilitative Carriers (SLC19A1-3 and SLC46A1) and Folate Receptors.’, Molecular Aspects of Medicine, 2013;34: 373–385
- Tang, Y.-S., R A Khan, Y Zhang, S Xiao, M Wang, D K Hansen, and others, ‘Incrimination of Heterogeneous Nuclear Ribonucleoprotein E1 (HnRNP-E1) as a Candidate Sensor of Physiological Folate Deficiency’, Journal of Biological Chemistry, 2011;286: 39100–39115
- Wang, Xiaoxia, Meijuan Chen, Jing Zhou, and Xu Zhang, ‘HSP27, 70 and 90, Anti-Apoptotic Proteins, in Clinical Cancer Therapy (Review).’, International Journal of Oncology, 2014;45: 18–30
- Toffoli, G, C Cernigoi, A Russo, A Gallo, M Bagnoli, and M Boiocchi, ‘Overexpression of Folate Binding Protein in Ovarian Cancers.’, International Journal of Cancer, 1997; 74: 193–198
- Kalli, Kimberly R, Ann L Oberg, Gary L Keeney, Teresa J H Christianson, Philip S Low, Keith L Knutson, and others, ‘Folate Receptor Alpha as a Tumor Target in Epithelial Ovarian Cancer.’, Gynecologic Oncology, 2008; 108: 619–626
- O’Shannessy, Daniel J, Gordon Yu, Robert Smale, Yao-Shi Fu, Sunil Singhal, Robert P Thiel, and others, ‘Folate Receptor Alpha Expression in Lung Cancer: Diagnostic and Prognostic Significance.’, Oncotarget, 2012; 3:414–25
- Nunez, Maria Ines, Carmen Behrens, Denise M Woods, Heather Lin, Milind Suraokar, Humam Kadara, and others, ‘High Expression of Folate Receptor Alpha in Lung Cancer Correlates with Adenocarcinoma Histology and EGFR [Corrected] Mutation.’, Journal of Thoracic Oncology : Official Publication of the International Association for the Study of Lung Cancer, 2012; 7: 833–840
- Hartmann, Lynn C, Gary L Keeney, Wilma L Lingle, Teresa J H Christianson, Bindu Varghese, David Hillman, and others, ‘Folate Receptor Overexpression Is Associated with Poor Outcome in Breast Cancer.’, International Journal of Cancer, 2007; 121: 938–942
- Chen, Kang, Li Xiong, Zhuling Yang, Shengfu Huang, Rong Zeng, and Xiongying Miao, ‘Prothymosin-Alpha and Parathymosin Expression Predicts Poor Prognosis in Squamous and Adenosquamous Carcinomas of the Gallbladder.’, Oncology Letters, 2018; 15: 4485–4494
- Hannappel, Ewald, and Thomas Huff, ‘The Thymosins. Prothymosin Alpha, Parathymosin, and Beta-Thymosins: Structure and Function.’, Vitamins and Hormones, 2003;66: 257–296
- Wang, M, J-Y Pan, G-R Song, H-B Chen, L-J An, and S-X Qu, ‘Altered Expression of Estrogen Receptor Alpha and Beta in Advanced Gastric Adenocarcinoma: Correlation with Prothymosin Alpha and Clinicopathological Parameters.’, European Journal of Surgical Oncology : The Journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology, 2007; 33: 195–201
- Jou, Yeong-Chin, Chun-Liang Tung, Yuh-Shyan Tsai, Cheng-Huang Shen, Chen Syue-Yi, Ai-Li Shiau, and others, ‘Prognostic Relevance of Prothymosin-Alpha Expression in Human Upper Urinary Tract Transitional Cell Carcinoma.’, Urology, 2009; 74:951–957
- Ha, Sang Yun, Dae Hyun Song, Soo Hyun Hwang, Soo Youn Cho, and Cheol-Keun Park, ‘Expression of Prothymosin Alpha Predicts Early Recurrence and Poor Prognosis of Hepatocellular Carcinoma.’, Hepatobiliary & Pancreatic Diseases International : HBPD INT, 2015; 14:171–177








