Karthikeyan S.1 , Sanjay Kumar P.1
, Sanjay Kumar P.1 , R J Madhusudan2, S K Sundaramoorthy2, P K Krishnan Namboori3*
, R J Madhusudan2, S K Sundaramoorthy2, P K Krishnan Namboori3*
1Amrita Molecular Modeling and Synthesis (AMMAS) Research lab, Center for Computational Engineering and Networking (CEN), Amrita School of Engineering, Coimbatore, Amrita Vishwa Vidyapeetham, India.
2Lotus Eye Hospital and Institute, Avinashi Road, Coimbatore-641 014
3Amrita Molecular Modeling and Synthesis (AMMAS) Research lab, Center for Computational Engineering and Networking (CEN), Amrita School of Engineering, Coimbatore, Amrita Vishwa Vidyapeetham, India.
Corresponding Author E-mail: n_krishnan@cb.amrita.edu
DOI : https://dx.doi.org/10.13005/bpj/1788
Abstract
The health-related complications such as diabetes, macular degeneration, inflammatory conditions, ageing and fungal infections may cause damages to the retina and the macula of the eye, leading to permanent vision loss. The major diseases associated with retina are Arteriosclerotic retinopathy (AR), Central retinal vein occlusion (CRVO), Branch retinal artery occlusion (BRAO), Coat's disease (CD) and Hemi-Central Retinal Vein Occlusion (HRVO). The symptomatic variations among these disorders are relatively confusing so that a systematic diagnostic strategy is difficult to set in. Therefore, an early detection device is required that is capable of differentiating the various ophthalmic complications and thereby helping in providing the right treatment to the patient at the right time. In this research work, 'Deep Convolution Neural Networks (Deep CNN) based machine learning approach has been used for the detection of the twelve major retinal complications from the minimal set of fundus images. The model was further cross-validated with real-time fundus images. The model is found to be superior in its efficiency, specificity and ability to minimize the misclassification. The “multi-class retinal disease” model on further cross-validation with real-time fundus image of the gave an accuracy of 95.63 %, validation accuracy of 92.99 % and F1 score of 91.96 %. The multi-class model is found to be a theranostic clinical support system for the ophthalmologist for diagnosing different kinds of retinal problems, especially BRAO, BRVO, CRAO, CD, DR, HRVO, HP, HR, and CN.
Keywords
Deep CNN; Fundus Image; Multi-Class Retinal Diseases; Theranostic Tool
Download this article as:| Copy the following to cite this article: Karthikeyan S., Kumar P. S, Madhusudan R. J, Sundaramoorthy S. K, Namboori P. K. K. Detection of Multi-Class Retinal Diseases Using Artificial Intelligence: An Expeditious Learning Using Deep CNN with Minimal Data. Biomed Pharmacol J 2019;12(3). |
| Copy the following to cite this URL: Karthikeyan S., Kumar P. S, Madhusudan R. J, Sundaramoorthy S. K, Namboori P. K. K. Detection of Multi-Class Retinal Diseases Using Artificial Intelligence: An Expeditious Learning Using Deep CNN with Minimal Data. Biomed Pharmacol J 2019;12(3). Available from: https://bit.ly/2oTcpvU |
Introduction
There are a number of retinal diseases reported so far such as Arteriosclerotic retinopathy (AR), Central retinal vein occlusion (CRVO), Central retinal artery occlusion (CRAO), Branch retinal vein occlusion (BRVO), Branch retinal artery occlusion (BRAO), Coat’s disease (CD), Hemi-Central Retinal Vein Occlusion (HRVO), Histoplasmosis (HP), Hypertensive retinopathy (HR), Choroidal neovascularization (CNV) and diabetic retinopathy (DR), which may even lead to permanent vision loss. Out of these, age-related macular degeneration (AMD) and diabetic retinopathy have been identified as the most significant.1, 2
The restrictions of the human eye and inadequacy of the conventional techniques to diagnose the various types of retinal diseases accurately and early in advance are the major challenges faced by the present-day ophthalmologists in the correct treatment of the patients.2-4 At present, highly sophisticated and dependable diagnostic imaging techniques such as ‘Fluorescent Retinal Angiography (FRA) and Optical Coherence Tomography (OCT) etc. are very popular. A countless number of machine learning approaches such as the artificial neural network (ANN), K-nearest neighbor algorithm, support vector machine (SVM) and Naive Bayes classifier (NBC) are incorporated to improve the prediction accuracy towards the detection of retinal diseases from the fundus images. It has been reported that ‘ANN based algorithms’ are efficient in predicting glaucoma from fundus images.5-8
Traditional classification approaches depend on feature extraction and feature classification techniques designed for the specific problem based on the available knowledge of the field. Most of the algorithms used in this area encounter the challenge of having the only insufficient number of datasets for training the model through conventional machine learning techniques.9 The launching of ‘deep learning CNN’ based algorithms makes evolutionary changes in the approach by directly identifying features from the training data without the categorical elaboration on feature extraction and classification. However, deep learning-based models are found to improve their efficiency on vigorous training using a large number of datasets.10 The availability of medical images will be highly limited in most cases, causing difficulties in using deep learning-based algorithms in the field of healthcare.8, 9 A number of open platforms and databases have been developed to store healthcare related medical images around the world. The techniques based on ‘deep extraction of information from the available images’ such as ‘affine transformation’ and ‘rotation of the images’ have been introduced to improve the efficiency of deep learning-based algorithms.7- 10
The deep learning based prediction strategy can be extensively used in diabetic retinopathy (DR).9 An unconventional and evolutionary deep learning prediction model for diagnosing DR by an automatic feature extraction learning method has been developed by Google, which helps even to grade and classify the intensity of nuclear cataract among the patients.7-10 Similarly, many deep learning approaches have been introduced in the prediction of ‘Retinopathy of prematurity (ROP)’ and AMD. Recent studies on fundus images using deep learning algorithms establish the possibility of predicting cardiovascular risk factors, suggesting these images as potential predictive models. Due to the lack of the accessible patient database, most of the predictive models designed so far have been set only for binary classification or to identify the presence of only diabetic retinopathy.9-11
The possibility of using fundus images for the design of ‘multi categorical predictive model’ covering various retinal diseases has been tried in the present work. The designed model is used to diagnose and differentiate retinal diseases such as arteriosclerotic retinopathy, branch retinal vein occlusion, etc.
Methods
Data Collection
The fundus images corresponding to different retinal disease were acquired from three standard online databases namely, ‘DIARETDB0, HRF Image Database and STARE’ for the primary training of the model. Similarly, 91 real-time images were acquired and clinically diagnosed and categorized by practising ophthalmologists, for training and cross-validation.
A total of 130 images were acquired from DIARETDB0, (110 are of diabetic retinopathy – haemorrhages, soft exudates, hard exudates, neovascularization and micro aneurysm] and 20 are from normal healthy people.12 From ‘High-Resolution Fundus (HRF) Image Database’ 15 images (normal, DR and glaucoma) were obtained.13 The images have been collected for the twelve major classes of diseases such as AR, CRVO, CRAO, BRVO, BRAO, CD, HRVO, HP, HR, CN, DR and normal retina. Additionally, the prediction models have been trained efficiently with minimal retinal images and have been cross-validated using real-time fundus image of patients from the hospital.
Similarly, from the ‘Structured Analysis of the Retina (STARE) online database, 14 categories of 397 images including ‘Background Diabetic Retinopathy (BDR)’, ‘Proliferative Diabetic Retinopathy (PDR)’, retinitis, emboli, CRAO, CRVO, BRAO, etc. were collected.14
The images were collected from various databases to improve the availability of various classes for analysis. The images were rotated to different angles and the corresponding data were also included in the samples to be tested leading to an increase in the effective number of images to 2484. Out of these images, 80% has been used for training and the remaining 20% for testing. The images were then resized to 224 x 224 pixels for optimum image resolution15- 18 the detailed information regarding data collection and dataset preparation has been included in Table 1.
Table 1: Detailed Information Regarding Data Collection
| Sl. No | Name of the disease | DIRETDB0 | HRF retinal database | STARE | Real time images |
| 1 | Arteriosclerotic retinopathy [AR] | 0 | 0 | 20 | 0 |
| 2 | Central retinal vein occlusion [CRVO] | 0 | 0 | 25 | 14 |
| 3 | Central retinal artery occlusion [CRAO] | 0 | 0 | 8 | 9 |
| 4 | Branch retinal vein occlusion [BRVO] | 0 | 0 | 10 | 25 |
| 5 | Branch retinal artery occlusion [BRAO] | 0 | 0 | 5 | 0 |
| 6 | Coat’s disease [CD] | 0 | 0 | 10 | 0 |
| 7 | Hemi-Central Retinal Vein Occlusion [HRVO] | 0 | 0 | 10 | 0 |
| 8 | Histoplasmosis [HP] | 0 | 0 | 10 | 0 |
| 9 | Hypertensive retinopathy [HR] | 0 | 0 | 20 | 16 |
| 10 | Choroidal neovascularization [CN] | 0 | 0 | 50 | 27 |
| 11 | diabetic retinopathy[DR] | 110 | 15 | 75 | 0 |
| 12 | Normal | 20 | 15 | 36 | 0 |
Deep Learning Architecture
The deep predictive model has been designed based on a pre-trained model, which was developed by Oxford Visual Geometry Group [VGG] known as VGG19. The VGG pre-trained model is built on a 3×3 convolutional layers stacked up together to increase the depth, followed by a max-pooling layer to reduce the volume size. After convolution, these features are more readily learned by a fully connected neural network of 4,096 nodes.19 The learning weights have been optimized to achieve maximum accuracy. The model architecture used for the research has been shown in [Fig. 1].
 |
Figure 1: Deep CNN architecture |
Model Optimization
Initially, the number of the epoch was optimized as 50 to provide proper convergence and maximum accuracy. A loss function or scoring function permits the system to compute the efficiency of the classification. In addition, a categorical cross entropy loss function has been used for the evaluation of the VGG19-softmax model by predicting the accuracy and validation accuracy.20-22 Besides computing accuracy, sensitivity, specificity, and precision have been taken into account and a confusion matrix has been generated for calculating the true positive rate (TPR) and false positive rate (FPR), which reflect the detailed performance information of the classifier.21- 25
Results and Discussion
The prediction model gave an accuracy of 95.63 % and validation accuracy of 92.99 %, supporting its effectiveness in the prediction from the image samples [Fig. 2].
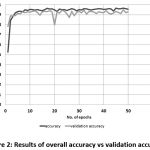 |
Figure 2: Results of overall accuracy vs validation accuracy |
A confusion matrix has been computed for further evaluation of the classifiers [Table 2]. It has been found that the model predicts most of the diseases accurately especially, BRAO, BRVO, CRAO, CD, DR, HRVO, HP, HR, and CN. However, the prediction accuracy of the model to the diseases AR and CRVO from the fundus images collected was very low [Fig. 3].
Table 2: Confusion Matrix Result of Our Model
| N=499 | AR | BRAO | BRVO | CRAO | CRVO | CD | DR | HRVO | HP | HR | CN | Normal | |
| AR | 15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 |
| BRAO | 0 | 35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 35 |
| BRVO | 0 | 0 | 34 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 35 |
| CRAO | 7 | 0 | 0 | 32 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 40 |
| CRVO | 0 | 0 | 0 | 0 | 41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 41 |
| CD | 0 | 0 | 0 | 0 | 0 | 33 | 0 | 0 | 0 | 0 | 0 | 0 | 33 |
| DR | 5 | 0 | 0 | 1 | 0 | 0 | 54 | 0 | 0 | 0 | 1 | 0 | 61 |
| HRVO | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 0 | 0 | 0 | 0 | 40 |
| HP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 0 | 0 | 0 | 40 |
| HR | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 56 | 1 | 0 | 62 |
| CN | 0 | 0 | 0 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 48 | 0 | 53 |
| Normal | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 41 |
| 35 | 38 | 34 | 36 | 42 | 36 | 56 | 40 | 40 | 56 | 50 | 36 |
 |
Figure 3: Accuracy results of all the categories value of our VGG19-softmax model. |
The true positive rate (TPR), a measure of sensitivity has been plotted out of the confusion matrix as shown in [Fig. 4].
 |
Figure 4: TPR results of all the categories value of our VGG19-softmax model. |
The sensitivity score of all the diseases was found to be high, excepting AR. In a few cases such as BRVO, HR, HRVO, HP and Normal, the sensitivity was 100%. Moreover, the false positive rate of these classes is zero [Fig. 5].
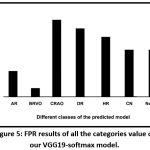 |
Figure 5: FPR results of all the categories value of our VGG19-softmax model. |
The precision values of these classes are found to be high [Fig. 6] suggesting a low level of misclassification. The specificity, which is a measure of the percentage of negatives that have been correctly identified, for the “multi-class retinal disease prediction model” has been found to be greater than 95%, supporting minimum misclassification [Fig. 7]. F1-score of the classes has been shown in [Fig. 8].
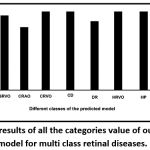 |
Figure 6: Precision results of all the categories value of our VGG19-softmax model for multi class retinal diseases. |
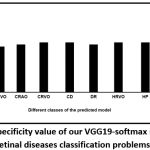 |
Figure 7: Results of specificity value of our VGG19-softmax model for multi-class retinal diseases classification problems. |
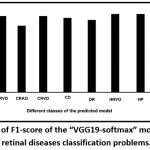 |
Figure 8: Results of F1-score of the “VGG19-softmax” model for multiclass retinal diseases classification problems. |
All the parameters characterizing the prediction model are included in [Table 3]. The designed prediction tool is found to be a ‘potential multi-class retinal disease prediction model’ from fundus images.
Table 3: results of all the parameters from confusion matrix
| Name of the classes | Accuracy | Precision | TPR | Specificity | FPR | F1-score |
| AR | 95.28 | 83.33 | 42.86 | 99.34 | 0.66 | 56.60 |
| BRAO | 99.36 | 100.00 | 92.11 | 100.00 | 0.00 | 95.89 |
| BRVO | 99.78 | 97.14 | 100.00 | 99.77 | 0.23 | 98.55 |
| CRAO | 97.48 | 80.00 | 88.89 | 98.18 | 1.82 | 84.21 |
| CRVO | 99.78 | 100.00 | 97.62 | 100.00 | 0.00 | 98.80 |
| CD | 99.36 | 100.00 | 91.67 | 100.00 | 0.00 | 95.65 |
| DR | 98.10 | 88.52 | 96.43 | 98.32 | 1.68 | 92.31 |
| HRVO | 100.00 | 100.00 | 100.00 | 100.00 | 0.00 | 100.00 |
| HP | 100.00 | 100.00 | 100.00 | 100.00 | 0.00 | 100.00 |
| HR | 98.72 | 90.32 | 100.00 | 98.55 | 1.45 | 94.92 |
| CN | 98.51 | 90.57 | 96.00 | 98.81 | 1.19 | 93.20 |
| Normal | 98.93 | 87.80 | 100.00 | 98.85 | 1.15 | 93.51 |
With recent advancements in the field of medical image processing using machine learning algorithms and data mining techniques, the computer-aided medical diagnostics has become an inevitable part of healthcare. In deep learning platform, the binary class-based classifiers have shown better accuracy and performance than multiclass classifiers. The most common drawback faced by multiclass classifiers is the chance for over fitting. For the designed model, the validation accuracy is in close proximity to the training data, therefore the chances of the model to over fit is negligibly small. The lower values of FPR, the higher values of precision and specificity ensure the model to be more dependable with minimum chances for Misclassification.
Conclusion
The ‘multi-class retinal disease prediction model’ is found to be a potential device in predicting retinal diseases from the fundus images. The overall efficiency of the model is found to be 92%. This study supports the possibility of exploiting the opportunities of pre-trained models with different medical applications using deep learning techniques. The model seems to predict the diseases BRAO, BRVO, CRAO, CD, DR, HRVO, HP, HR, and CN.
The model could further be used to make individual variations associated with the images and various mutations corresponding to retinal diseases thereby making fundus image as a potential biomarker for various associated diseases like cardiovascular complications, Alzheimer’s disease, hypoxic conditions and etc.
Acknowledgments
The authors Karthikeyan S and Sanjay Kumar P express their gratitude to ‘Coconut Development Board’, Government of India for the financial support as fellowship.
Funding Source
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
The authors declare that there is no conflict of interest associated with the manuscript.
References
- Wiedemann P. Growth factors in retinal diseases: Proliferative vitreoretinopathy, proliferative diabetic retinopathy, and retinal degeneration. Surv Ophthalmol. 1992;36(5):373-384. doi:10.1016/0039-6257(92)90115-A
- Pardue MT, Allen RS. Neuroprotective strategies for retinal disease. Prog Retin Eye Res. 2018;65:50-76. doi:10.1016/j.preteyeres.2018.02.002
- Kermany DS, Goldbaum M, Cai W, et al., Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell. 2018;172(5):1122-1131.e9. doi:10.1016/j.cell.2018.02.010
- Valverde C, Garcia M, Hornero R, Lopez-Galvez M. Automated detection of diabetic retinopathy in retinal images. Indian J Ophthalmol. 2016;64(1):26. doi:10.4103/0301-4738.178140
- Ravudu M, Jain V, Kunda MMR. Review of image processing techniques for automatic detection of eye diseases. In: IEEE; 2012:320-325. doi:10.1109/ICSensT.2012.6461695
- Stella Mary MCV, Rajsingh EB, Naik GR. Retinal Fundus Image Analysis for Diagnosis of Glaucoma: A Comprehensive Survey. IEEE Access. 2016;4:4327-4354. doi:10.1109/ACCESS.2016.2596761
- Araújo T, Aresta G, Castro E, et al. Classification of breast cancer histology images using Convolutional Neural Networks. Sapino A, ed. PLOS ONE. 2017;12(6):e0177544. doi:10.1371/journal.pone.0177544
- Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316(22):2402. doi:10.1001/jama.2016.17216
- Li Y-H, Yeh N-N, Chen S-J, Chung Y-C. Computer-Assisted Diagnosis for Diabetic Retinopathy Based on Fundus Images Using Deep Convolutional Neural Network. Mob Inf Syst. 2019;2019:1-14. doi:10.1155/2019/6142839
- S K S, P A. A Machine Learning Ensemble Classifier for Early Prediction of Diabetic Retinopathy. J Med Syst. 2017;41(12). doi:10.1007/s10916-017-0853-x
- Choi JY, Yoo TK, Seo JG, Kwak J, Um TT, Rim TH. Multi-categorical deep learning neural network to classify retinal images: A pilot study employing small database. Liu B, ed. PLOS ONE. 2017;12(11):e0187336. doi:10.1371/journal.pone.0187336
- K DURAISWAMY MT. Automatic detection of microaneurysms using microstructure and wavelet methods. 2015;40(Part 4):1185–1203.
- Kolar R, Kubena T, Cernosek P, et al. Retinal vessel segmentation by improved matched filtering: evaluation on a new high-resolution fundus image database. IET Image Process. 2013;7(4):373-383. doi:10.1049/iet-ipr.2012.0455
- Hoover A, Goldbaum M. Locating the optic nerve in a retinal image using the fuzzy convergence of the blood vessels. IEEE Trans Med Imaging. 2003;22(8):951-958. doi:10.1109/TMI.2003.815900
- Sahebrao R, N. S, T. S, Dhopeshwarkar M. Automated Diagnosis Non-proliferative Diabetic Retinopathy in Fundus Images using Support Vector Machine. Int J Comput Appl. 2015;125(15):7-10. doi:10.5120/ijca2015905968
- Budai A, Bock R, Maier A, Hornegger J, Michelson G. Robust Vessel Segmentation in Fundus Images. Int J Biomed Imaging. 2013;2013:1-11. doi:10.1155/2013/154860
- Siva Sundhara Raja D, Vasuki S. Automatic Detection of Blood Vessels in Retinal Images for Diabetic Retinopathy Diagnosis. Comput Math Methods Med. 2015;2015:1-12. doi:10.1155/2015/419279
- Lam C, Yu C, Huang L, Rubin D. Retinal Lesion Detection With Deep Learning Using Image Patches. Investig Opthalmology Vis Sci. 2018;59(1):590. doi:10.1167/iovs.17-22721
- Das S, Malathy C. Survey on diagnosis of diseases from retinal images. J Phys Conf Ser. 2018;1000:012053. doi:10.1088/1742-6596/1000/1/012053
- Yalcin N, Alver S, Uluhatun N. Classification of retinal images with deep learning for early detection of diabetic retinopathy disease. In: IEEE; 2018:1-4. doi:10.1109/SIU.2018.8404369
- Landgrebe TCW, Duin RPW. Efficient Multiclass ROC Approximation by Decomposition via Confusion Matrix Perturbation Analysis. IEEE Trans Pattern Anal Mach Intell. 2008;30(5):810-822. doi:10.1109/TPAMI.2007.70740
- Razavian AS, Azizpour H, Sullivan J, Carlsson S. CNN Features off-the-shelf: an Astounding Baseline for Recognition. March 2014. http://arxiv.org/abs/1403.6382. Accessed May 24, 2019.
- Sanjay KUMAR P. Prediction of epigenetic variations in alzheimer’s disease identification of ethnic variants through pharmacogenomic approach. Res J Pharm Biol Chem Sci. 2016;7(4):2742-2745.
- A M H, C LA, O M D, Namboori PKK. Evaluation of Colorectal Cancer (CRC) Epidemiology A Pharmacogenomic Approach. J Young Pharm. 2017;9(1):36-39. doi:10.5530/jyp.2017.9.7
- Iyer PM, Karthikeyan S, Sanjay Kumar P, Krishnan Namboori PK. Comprehensive strategy for the design of precision drugs and identification of genetic signature behind proneness of the disease—a pharmacogenomic approach. Funct Integr Genomics. 2017;17(4):375-385. doi:10.1007/s10142-017-0559-7








