Nirmaladevi Ponnusamy , Rajasree Odumpatta, Pavithra Damodharan and Mohanapriya Arumugam*
, Rajasree Odumpatta, Pavithra Damodharan and Mohanapriya Arumugam*
Department of Biotechnology, Vellore Institute of Technology, Vellore, 632014-India.
Corresponding Author E-mail: mohanapriyaa@vit.ac.in
DOI : https://dx.doi.org/10.13005/bpj/1757
Abstract
In the present study, in silico analysis was employed to identify the action of marine bioactive compounds against KSHV targets. Virulence factor analysis of KSHV from literature review, three proteins LANA1, vIRF3/LANA2 and PF-8 were identified as putative drug targets. The quality of protein structures play a significant role in the experimental structure validation and prediction, where the predicted structures may contain considerable errors was checked by SAVES v5.0 servers. By virtual screening four potential bioactive compounds Ascorbic acid, Salicylihalamide A, Salicylihalamide B and Frigocyclinone were predicted. One of the potential compounds of Frigocyclinone has acting against KSHV proteins. Hence, determined as the good lead molecule against KSHV. Molecular dynamic simulation studies revealed the stability of LANA1- Frigocyclinone complex and it could be a futuristic perspective chemical compound for Kaposi’s sarcoma.
Keywords
Anti-Tumor Property and Angucyclinone Derivatives; Binding Energy; Bioactive Compounds; Frigocyclinone; KSHV
Download this article as:| Copy the following to cite this article: Ponnusamy N, Odumpatta R, Damodharan P, Arumugam M. Computational Investigation of Marine Bioactive Compounds Reveals Frigocyclinone as a Potent Inhibitor of Kaposi’s Sarcoma Associated Herpesvirus (KSHV) Targets. Biomed Pharmacol J 2019;12(3). |
| Copy the following to cite this URL: Ponnusamy N, Odumpatta R, Damodharan P, Arumugam M. Computational Investigation of Marine Bioactive Compounds Reveals Frigocyclinone as a Potent Inhibitor of Kaposi’s Sarcoma Associated Herpesvirus (KSHV) Targets. Biomed Pharmacol J 2019;12(3). Available from: https://bit.ly/2No5jJN |
Introduction
KSHV also called as Human herpesvirus 8 causes frequent vascular tumor most commonly seen in AIDS and immunosuppressed patients.1 Etiological agent of endothelium derived malignancy KS, primary effusion lymphoma, multicentric Castleman’s disease and germinotropic lympho-proliferative disorder are associated with KSHV. During KS pathogenesis, KSHV induced by COX-2 which regulates multiple events such as pro-inflammatory cytokines, growth factors, angiogenic factors, anti-inflammatory cytokines, matrix metalloproteinases and tissue inhibitors of metalloproteinases.2,3 KSHV reveals a biphasic cycle of lifelong rescindable latent phase and transient lytic reactivation phase, which has effectively distinctive gene expression outlines.4 However, inappropriate induction of lytic gene expression by reactivation stage indicates increased inflammatory cytokine levels (IL-1ß, TNFα, IL-6, IL-15 and IL-17) in blood and tissues with KS.5 Moreover pro-inflammatory cytokines (IL-1α, IL-1ß and IL-6) induce phenotypic and functional features in KSHV infection during KS histogenesis. Expression of anti-inflammatory cytokine responses (IL-4, IL-13 and IL-15) controls inflammation within epidermal units which is mainly initiated during latent phase by alpha-melanocortin stimulating hormone but fails to maintain lytic replication.6 KSHV genome with restricted region is transcriptionally active throughout latency, encrypts four main ORFs containing Latency-associated nuclear antigen or LANA1, viral-cyclin, viral FLICE-inhibitory protein, and Kaposins along with 18 mature miRNAs and viral interferon regulatory factor-3.7 The viral protein of LANA1 plays a vital role in modulating viral and cellular gene expression. LANA1 is enhancing the activity of the HIV-1 promoter via linked with Tat, and recognized virus encrypted as transactivator.8 ORF59 protein as PF-8 and that is presenting an early stage of lytic phase.9 PF-8 encrypts DNA polymerase and also homologous to express other herpesvirus such as HSV-1 UL42, Epstein-Barr virus, BMRF1, herpesvirus saimiri ORF59 protein, human cytomegalovirus ICP36, HHV-6 p41, varicella-zoster virus gene 16 protein, and HHV-7 U27. vIRF-3 is also known as LANA2 which influences B cells only10 (latent phase).
The bioactive compounds are derived from marine organisms. More than 30,000 bioactive compounds distinguished from various marine micro-organisms are shown to possess anti-bacterial, anti-inflammatory and also anti-tumor properties.11 The marine organisms like bacteria, sponge and micro-algae had a significant role in the pharmaceutical industry. One of the important marine red sponges of haliclona sp. produce alkaloids, macrolides, peptides, polyketides, polyacetylenes, steroids and halogenated derivatives as bioactive compounds. The halicona sp. produces salicylihalamide A and salicylihalamide B which comes under same family and species, whereas structurally and functionally different. These compounds have anti-tumor properties. Ascophyllum nodosum is a large brown algae, which belongs to the Phaeophyceae family and it is the only species in the genus Ascophyllum which produce bioactive compounds with anti-oxidant and immunostimulatory properties. Ascorbic acid is present in all red, brown, and green seaweeds that reduces the risk of cancer, cardiovascular and Alzheimer’s disease. Frigocyclinone isolated from Streptomyces griseusstrain NTK 97 possess a significant role in antibacterial and antitumor activities.
Our study focuses on identifying bioactive compounds from different marine organisms against kaposi’s sarcoma associated herpesvirus proteins.
Materials and Methods
Target Preparation
The X-ray crystal structures of the two proteins – LANA1 (PDB ID: 5A76),12 PF-8 (PDB ID: 3HSL)13 were retrieved from RCSB Protein Data Bank. The 3D structure of vIRF3 protein is not available in the Protein Data Bank. Therefore, the three-dimensional structure was build using homology modelling.
Homology Modelling of vIRF3
The vIRF3 (UniProtKB: F5HIC6) protein sequence was retrieved from Universal Protein Resource (http://www.uniprot.org/). Using BLASTP, the suitable template sequence was retrieved to identify the homologous structure. Hence, the 3D structure of vIRF3 protein was build using Modeller14 version 9.18, Swiss Modeller15 and ModWeb.16 The best protein model was chosen on the basis of the percentage identity and E value.
Ligand Preparation
A set of seventy bioactive compounds from marine organisms were collected from scientific literature. The two dimensional structure of all the compounds were retrieved from Pubchem database. The structure of ligands was converted from SDF to SMILE using Openbabel software.17 The purpose of virtual screening to find out the potential lead compounds with active function and high inhibitory activity against KSHV. The molecular properties (logP, polar surface area, number of hydrogen bond donors and acceptors and others), bioactivity score (GPCR ligands, kinase inhibitors, ion channel modulators, nuclear receptors) and drug likeness score were calculated by Molinspiration18 and Molsoft.19 The bioactive compounds which obeys lipinski’s rule were taken for further studies.20 To optimize the 3D structure of bioactive compounds CORINA software were used.21
Model Validation and Energy Minimization
The conformational stability of modelled protein backbones were estimated via Ramachandran plot using RAMPAGE server which determines the dihedral angles ψ against φ of amino acid residues.22 Additionally, to validate our model we checked the packing conformational quality of the model using ProSA,23 ERRAT 24 and QMEAN.25 The crystal structures and the model were energy minimized to obtain lowest delta G value using Swiss-PDB Viewer.26
Molecular Dynamic Simulation for Protein-ligand Complex
Fluctuations and conformational changes were recognized via molecular dynamic (MD) simulation process for 20ns. Evaluation of RMSD, RMSF and gyration of both protein and protein-ligand complex were determined using Gromacs version 4.5.5. The topology file was generated via Gromos96 forcefield, whereas protein-ligand complex file, gromacs coordinate file and gromacs topology were prepared using PRODRG server. The solvation and ions were added and generated in the topology file. The solvated protein was energy minimized through a steepest descent algorithm. After minimizing energy the equilibration step was carried out to restraint the MD simulation. Further potential energy, temperature, pressure and density calculation were assessed.29
Results
Crystal Structure of KSHV Viral Proteins
KSHV produces major proteins LANA1 (Latent), vIRF3 (Latent in B cells; Lytic in endotelial cells) and PF-8 (Lytic) involved in various stages of development (Figure 1). Crystal structure of LANA1 and PF-8 were retrieved from PDB. The vIRF3 protein structure is not available in PDB. Hence, the homology model was build. The modeling of vIRF3 protein was done using a template as 4P55_A (Resolution: 2.50 Å).
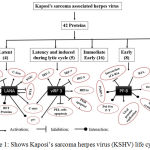 |
Figure 1: Shows Kaposi’s sarcoma herpes virus (KSHV) life cycle.
|
Construction of vIRF3 Protein Structure and Validation
The three dimensional structure of vIRF3 protein structure was constructed through various modeling methods such as Modeller 9.18 (Identity: 30%, E value: 0.002), Swiss model (Identity: 26%) and Modweb (Identity: 27%, E value: 0). The stereo chemical property of vIRF3 was evaluated and calculated through Ramachandran plot using the RAMPAGE server. The plot derives angle distribution of ψ and φ which is divided into three different regions. The plot reveals that homology model of vIRF3 contains 95% of residues in favored region and 5% of residues in allowed region; swiss model of vIRF3 contains 92% residues in favored region, 7% of residues in allowed region and 1% of residues in outer region; modweb model of vIRF3 contains 98% of residues in favored region and 2% of residues in allowed region.
An overall three dimensional quality of vIRF3 was measured by ProSA, ERRAT and QMEAN Z-Score. The Z score from ProSA server for all the three models are -3.77, -4.45 and -4.22 respectively. QMEAN Z-score of our model showed the range of values from -2.73 to -3.80. Though it deviates from the expected range of values from protein validation, still we considered the model since it showed better quality of structure with respect to Ramachandran plot.
Bioactive Compound Structural Identification
The present study mainly focuses to predict bioactive compounds against Kaposi’s sarcoma associated herpesvirus disease (Table 1). The chemical structure of compounds was obtained from PubChem database which were converted to three dimensional structure using a chemical toolbox, Openbabel. Virtual screening was implemented to retrieve the compounds that fit the Lipinski’s rule of five and possess drug-like properties. Compounds obeying Lipinski’s rule are further screened based on bioactivity and drug likeness score (Table 2(a), 2(b) & 2(c)). Out of seventy bioactive compounds, four bioactive compounds namely Ascorbic acid, Salicylihalamide A, Salicylihalamide B and Frigocyclinone showed good results.
Table 1: Marine source containing bioactive compounds with different species.
| S. No | Compound | Compounds family | Marine source producing species | Marine source family |
| 1 | Abyssomicin C30 | Polyketide | Verrucosispora sp. | Micromonosporaceae |
| 2 | Aeroplysinin-131 | Alkaloid | Verongia aerophoba | Sponge |
| 3 | Agar32 | Sulfated polysaacharide | Gracilaria dominguensis | Red algae |
| 4 | Alpha tocopherol33 | Tocopherol (vitamin E) | Ascophyllum Nodosum | Phaeophyceae (Brown algae) |
| 5 | Aplysiatoxin34 | Cyanotoxin | Lyngbya Majusula | Blue green algae |
| 6 | Ascididemin35 | Aromatic alkaloid | Didemnum sp. | Sponge |
| 7 | Ascorbic acid36 | Vitamin C | Ascophyllum Nodosum | Phaeophyceae (Brown algae) |
| 8 | Astaxanthin37 | Keto carotenoid | Haematococcus pluvialis | Chlorophyta (Green algae) |
| 9 | Aureoverticillactam38 | Macrocyclic lactam | Streptomyces aureoverticillatus | Bacterium |
| 10 | Beta carotene39 | carotenoids | Dunaliella salina | Chlorophyta (Green algae) |
| 11 | Beta glucans40 | Polysaacharide | Laminaria Digitata | Laminariceae (Brown algae) |
| 12 | Caprolactones41 | Lactone | Streptomyces sp. | Streptomycetaceae |
| 13 | Chandrananimycins42 | Antibiotics | Actinomadura sp. | Thermomonosporaceae |
| 14 | Citrinadin A43 | Spirooxindole alkaloid | actinotrichia fragilis | Red algae |
| 15 | Curacin A44 | Thiazole lipid | Lyngbya majuscula | Cyanobacterium |
| 16 | Desmosterol45 | Sterols | Palmaria species Porphyra sp. | Red algae |
| 17 | Dictyodendrins46 | Pyrrolocarbazole derivatives | Dictyodendrilla verongiformis | Sponge |
| 18 | Dictyol C47 | Diterpenes | Dictyota dichotoma | Brown algae |
| 19 | Dictyol H48 | Diterpenes | Dictyota dentate | Brown algae |
| 20 | Dicurcuphenol A49 | Sesquiterpene | Didiscus aceratus | Sponge |
| 21 | Discodermolide50 | Lactone | Discodermia dissoluta | Sponge |
| 22 | DMMC51*1 | Cyclic depsipeptide | Lyngbya majuscula | Cyanobacterium |
| 23 | Docosahexaenoic acid52 | PUFA*2 | Schizochytrium sp. | Marine Microalgae |
| 24 | Dolabellanes53 | Diterpenes | Dilophus spiralis | Dictyotaceae (Brown algae) |
| 25 | Dominicin54 | Octapeptide | Eurypon laughlini | Caribbean sponge |
| 26 | Halichondrin B55 | Macro cyclic polyether | Halichondria okadai | Sponge |
| 27 | Eicosapentaenoic acid56 | PUFA*2 | Monodus subterraneus | Marine Microalgae |
| 28 | Spisulosine (ES-285)57 | Alkyl amino alcohol | Mactromeris polynyma | Mollusc |
| 29 | Frigocyclinone58 | Angucyclinone antibiotic | Streptomyces griseus | Bacterium |
| 30 | Fucoidan59 | Sulfated polysaccharide | Fucus vesiculosus | Brown algae |
| 31 | Fucosterol60 | Sterols | Laminaria ochroleuca Undaria pinnatifida | Brown algae |
| 32 | Fucoxanthin61 | carotenoid | Fucus vesiculosus | Brown macro-algae |
| 33 | Glaciapyrroles62 | pyrrolosesquiterpenes | Streptomyces sp. | Streptomycetaceae |
| 34 | Griffithsin63 | Lectin (protein) | Griffithsia | Red algae |
| 35 | Gutingimycin64 | polar trioxacarcin | Streptomyces sp. | Streptomycetaceae |
| 36 | Helquinoline65 | Tetrahydroquinoline antibiotic | Janibacter limosus | Janibacter |
| 37 | Himalomycin A66 | Antibiotics | Streptomyces sp. | Streptomycetaceae |
| 38 | Himalomycin B66 | Antibiotics | Streptomyces sp. | Streptomycetaceae |
| 39 | Hemiasterlin (HTI-286)67 | Linear peptide | Cymbastella sp. | Sponge |
| 40 | Keramadine68 | Brominated alkaloid | Agelas sp. | Sponge |
| 41 | Komodoquinone A69 | Anthracycline | Streptomyces sp. | Streptomycetaceae |
| 42 | Bengamide B (LAF-389)70 | Ɛ-Lactam peptide derivative | Jaspis digonoxea | Sponge |
| 43 | Lajollamycin71 | Antibiotics | Streptomyces nodosus | Actinomycetes |
| 44 | Lambda carrageenan72 | Sulfated polysaacharide | Gigartina skottsbergii | Gigartinaceae (Red algae) |
| 45 | Lamellarin D73 | Pyrrole alkaloid | Lamellaria sp. | Mollusk |
| 46 | Laminarin74 | Polysaacharide | laminaria hyperborean | Brown seaweed |
| 47 | Laulimalide75 | Macrolide | Cacospongia mycofijiensis | Sponge |
| 48 | Laurebiphenyl76 | Sesquiterpene | Laurencia tristicha | Red algae |
| 49 | Lutein77 | carotenoids | Muriellopsis sp. | Chlorophycean (Green algae) |
| 50 | Marinomycins78 | Antibiotics | Marinispora | Actinomycete |
| 51 | Mechercharmycins79 | Cytotoxin | Thermoactinomyces sp. | Thermoactinomycetaceae |
| 52 | MKN-349A80 | Cyclic tetrapaptide | Nocardiopsis sp. | Nocardiopsaceae |
| 53 | Neopetrosiamide A81 | Linear peptide | Neopetrosia sp. | Sponge |
| 54 | Palythine82 | Mycosporine amino acid | Gelidium corneum | Red algae |
| 55 | Peloruside A83 | Macrocyclic lactone | Mycale hentscheli | Sponge |
| 56 | Phlorofucofuroeckol A84 | Phlorotannins | Ecklonia cava | Brown algae |
| 57 | Phlorofucofuroeckol B85 | Phlorotannins | Myagropsis myagroides | Sargassaceae (Brown seaweed) |
| 58 | Phlorotannins86 | Polyphenol | Sargassum fusiforme | Brown algae |
| 59 | Phycocyanobilins87 | Phycobiliproteins | Cyanobacteria, Rhodophyta | Blue green algae |
| 60 | Phycoerythrobilins88 | Phycobiliproteins | Rhodophyta | Red algae |
| 61 | Plakortone Q89 | Polyketide | Plakortis sp. | Sponge |
| 62 | Salicylihalimide A90 | Polyketide | Haliclona sp. | Sponge |
| 63 | Salicylihalimides B90 | Polyketide | Haliclona sp. | Sponge |
| 64 | Salinosporamide A91 | Bicyclic g-lactam-h lactone | Salinospora sp. | Actinomycete |
| 65 | Sarcodictyins92 | Diterpene | Sarcodictyon roseum | Coral |
| 66 | Shinorine83 | Mycosporine amino acid | Ahnfeltiopsis devoniensis | Red algae |
| 67 | Thiocoraline93 | Depsipeptide | Micromonospora marina | Actinomycete |
| 68 | Trioxacarcins94 | Antibiotics | Streptomyces sp. | Streptomycetaceae |
| 69 | Variolin B95 | Heterocyclic alkaloid | Kirkpatrickia variolosa | Sponge |
| 70 | Zeaxanthin96 | Carotenoid | Himanthalia Elongata | Brown seaweed |
*1 Desmethoxymajusculamide C, *2 Polyunsaturated fatty acid.
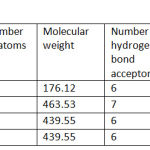 |
Table 2.a: Molecular properties of bioactive compounds.
|
Table 2.b: Bioactive score of bioactive compounds.
| S.No | Bioactive compounds | GPCR ligand | Ion channel modulator | Kinase inhibitor | Nuclear receptor ligand | Protease inhibitor | Enzyme inhibitor |
| 1 | Ascorbic acid | -0.53 | -0.24 | -1.09 | -1.01 | -0.81 | 0.20 |
| 2 | Frigocyclinone | 0.32 | 0.05 | 0.02 | 0.15 | 0.26 | 0.49 |
| 3 | Salicylihalamide A | 0.41 | 0.29 | 0.01 | 0.45 | 0.25 | 0.58 |
| 4 | Salicylihalamide B | 0.41 | 0.29 | 0.01 | 0.45 | 0.25 | 0.58 |
Table 2.c: Drug likeness score of bioactive compounds.
| S.No | Bioactive compounds | Druglikeness score |
| 1 | Ascorbic acid | 0.84 |
| 2 | Frigocyclinone | 0.93 |
| 3 | Salicylihalamide A | 1.01 |
| 4 | Salicylihalamide B | 1.01 |
Molecular Docking
Docking studies will help in appropriate consideration of the protein’s active site and its interaction with the ligand. The interaction between a small molecule and a protein may result in inhibition of the protein. Molecular docking program Autodock 4.2 was used in this study. The protein was energy minimized using Swiss PDB viewer. The result of the docking were analyzed based on the interactions and binding energies between KSHV proteins and the bioactive compounds. From the analysis, we found that frigocylinone has shown significant affinity towards LANA1 with binding energy of -8.59 kcal/mol followed by vIRF3 of -8.48 kcal/mol and with PF-8 of -8.00 kcal/mol. Among the three complexes, the LANA1-Frigocyclinone complex was known to possess better binding affinity with least binding energy (Table 3).
Table 3: Interacting of target-ligand energy values.
| S. No | Targets | Ascorbic acid (Kcal/mol) | Salicylihalamide A (Kcal/mol) | Salicylihalamide B (Kcal/mol) | Frigocyclinone (Kcal/mol) |
| 1 | LANA1 | -4.30 | -5.88 | -5.59 | -8.59 |
| 2 | vIRF3 | -5.45 | -8.42 | -8.06 | -8.48 |
| 3 | PF-8 | -4.75 | -5.36 | -6.09 | -8.00 |
The predicted results of LANA1-Frigocyclinone complex revealed best binding affinity, lowest binding energy of -8.59 Kcal/mol and formed H-bond with the residue LYS1070 (Table 4(a) & 4(b)). The results of docking studies indicates that the amino acid residues LYS1030, ALA1031, PRO1033, GLN1034, LYS1070, TRP1122, HIS1126, LEU1128 and ALA1129 play an important role in drug interaction. LYS1030, PRO1033, PHE1037, LYS1070, TRP1122, HIS1126 and LEU1128 were known to form hydrogen bonds with the compounds. The docking result shows that the amino acids LYS1070 and LEU1128 are involved in the interaction with more than one compound (Figure 2 & 3).
Table 4.a: Docked complex with residues and number of hydrogen bonds.
| S .No | Target-ligand complex | Residues | No. of hydrogen bonds | Binding energy (Kcal/mol) |
| 1 | LANA1 : Ascorbic acid | GLN1034, GLY1067, ARG1119, GLY1130 | 4 | -4.3 |
| 2 | LANA1 : Frigocyclinone | LYS1070 | 3 | -8.59 |
| 3 | LANA1 : Salicylihalamide A | LYS1070 | 2 | -5.88 |
| 4 | LANA1 : Salicylihalamide B | LYS1070 | 1 | -5.59 |
| 5 | vIRF 3 : Ascorbic acid | GLN51, ASP55, ARG58 | 6 | -5.45 |
| 6 | vIRF 3 : Frigocyclinone | ASN42 | 1 | -8.48 |
| 7 | vIRF 3 : Salicylihalamide A | ASN42 | 1 | -8.42 |
| 8 | vIRF 3 : Salicylihalamide B | ASN42, ASP43, GLN51, PHE53 | 4 | -8.06 |
| 9 | PF-8 : Ascorbic acid | GLU158, PHE153 | 3 | -4.75 |
| 10 | PF-8 : Frigocyclinone | HIS154, GLU158 | 2 | -8.00 |
| 11 | PF-8 : Salicylihalamide A | LYS63, SER288, GLY289 | 3 | -5.36 |
| 12 | PF-8 : Salicylihalamide B | LYS292, HIS154 | 3 | -6.09 |
Table 4.b: Different interaction values for major target-ligand complex measured through DSV.
| S. No | Target-ligand complex | Hydrogen bonds interaction | Electrostatic interaction | Hydrophobic interaction | Van der waals interaction | Miscellaneous | Unfavoured bump |
| 1 | LANA1 : Frigocyclinone | 3 | – | 6 | 5 | – | – |
| 2 | vIRF 3 : Frigocyclinone | 1 | – | 1 | 6 | 1 | – |
| 3 | PF-8 : Frigocyclinone | 2 | 2 | 4 | 10 | – | 1 |
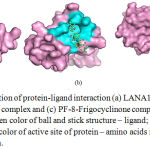 |
Figure 2: Pymol visualization of protein-ligand interaction (a) LANA1-Frigocyclinone complex, (b) vIRF3-Frigocyclinone complex and (c) PF-8-Frigocyclinone complex which represents different color such as green color of ball and stick structure – ligand; pink color of surface structure – protein; cyans color of active site of protein – amino acids residues; yellow – hydrogen bond interaction.
|
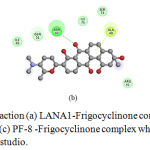 |
Figure 3: Protein-ligand interaction (a) LANA1-Frigocyclinone complex, (b) vIRF3 Frigocyclinone complex and (c) PF-8 -Frigocyclinone complex which visualized through discovery studio.
|
Therefore, the frigocyclinone with best inhibitory constant effect of 607.94 nM against LANA1 makes an intermolecular energy -8.89 Kcal/mol and electrostatic energy +0.04 kcal/mol. However, the complex possessed torsional energy value of +1.10 kcal/mol with the zero unbound energy and cluster RMSD 0.00 Å as well as reference RMSD 48.635 Å. The analysis of LANA1-Frigocyclinone complex hydrogen bond donor (LYS1070 (NZ) and acceptor (UNK25 (C4) & UNK31 (O16)) distance of 2.8Å & 2.9Å. and H-bond angle of ≥ 77º.
Molecular Dynamic Simulation of LANA1- Frigocyclinone Complex
To confirm docking analysis we did molecular dynamics simulation of LANA1-Frigocyclinone complex. We determined conformational changes between LANA1 and LANA1-Frigocyclinone complex. The results showed that LANA1-Frigocyclinone complex had average potential energy -270168 kJ/mol (total drift: -252845 kJ/mol), temperature 299.813 K (total drift: 1.01817 K), pressure -3.67818 bar (total drift: 12.6906 bar) and density 1005.7 kg/m3 (total drift: 0.35516 kg/m3). The steepest descents algorithm converged to Fmax<1000 in 1583 steps (potential energy: -5.3628425e+05). The LANA1 protein contains 1156 atoms and Frigocyclinone contains 37 atoms. RMSD curves indicate a slight changes between 8.93 ns and 8.96 ns whereas drastic increase relative to the docked conformation with values range between 10 ns and 20 ns. The LANA1-Frigocyclinone complexes produce more fluctuations during 3 ns and 13 ns. The radius of gyration were intended to determine the compactness of LANA1 during the MDS. All the position were compact with the LANA1 having the lowest Rg value of 1.31 nm at 16 ns and highest Rg value 1.37 nm at 4 ns (Figure 4).
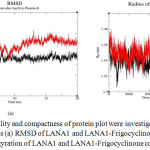 |
Figure 4: The stability and compactness of protein plot were investigated through MD simulations at 20 ns (a) RMSD of LANA1 and LANA1-Frigocyclinone complex and (b) Radius of gyration of LANA1 and LANA1-Frigocyclinone complex.
|
The LANA1 represents as black colour whereas LANA1-Frigocyclinone complex represents as red colour.
Conclusion
The current analysis, investigated the role of marine bioactive compounds as anticancer agents using computational methods. The result from this study displayed that the frigocyclinone demonstrated high affinity towards KSHV LANA1. Interaction analysis revealed that this compounds formed stable interaction in the surface of LANA1 mainly through H-bond. By this compound analysis, we provide a valuable insight on the identification of potent bioactive compound from marine source against KSHV. The main chemical component frigocyclinone is the first angucyclinone derivates (acts as antiviral, antifungal, anti-tumor and enzyme inhibitory activities). Therefore the compound frigocyclinone can be considered as promising anticancer lead for KS.
Acknowledgements
The authors acknowledge the management of Vellore Institute of Technology for providing the computer facilities and encouragement to this research work.
Conflict of interest
There is no conflict of interest.
References
- Beral V, Peterman T. A, Berkelman R. L and Jaffe H. W. Kaposi’s sarcoma among persons with AIDS: a sexually transmitted infection? Lancet., 1990; 335(8682): 123-128.
- Host K. M, Jacobs S. R, West J. A, Zhang Z, Costantini L. M, Stopford C. M, Dittmer D. P and Damania B. Kaposi’s Sarcoma-Associated Herpesvirus Increases PD-L1 and Proinflammatory Cytokine Expression in Human Monocytes. Imperiale MJ. ed. mBio., 2017; 8(5): e00917.
- Sharma-Walia N, Paul A. G, Bottero V, Sadagopan S, Veettil M. V, Kerur N and Chandran B. Kaposi’s Sarcoma Associated Herpes Virus (KSHV) Induced COX-2: A Key Factor in Latency, Inflammation, Angiogenesis, Cell Survival and Invasion. Nelson JA, ed. PLoS Pathogens., 2010; 6(2): e1000777.
- Dourmishev L. A, Dourmishev A. L, Palmeri D, Schwartz R. A and Lukac D. M. Molecular genetics of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol Mol. Biol. Rev., 2003; 67(2): 175-212.
- Stürzl M, Zietz C, Monini P and Ensoli B. Human herpesvirus-8 and Kaposi’s sarcoma: relationship with the multistep concept of tumorigenesis. Adv. Cancer Res., 2001; 81: 125-159.
- Fontana J. M, Mygatt J. G, Conant K. L, Parsons C. H and Kaleeba J. A. R. Kaposi’s Sarcoma-Associated Herpesvirus Subversion of the Anti-Inflammatory Response in Human Skin Cells Reveals Correlates of Latency and Disease Pathogenesis. J. Skin Cancer., 2014; 2014: 246076.
- Mesri E. A, Cesarman E and Boshoff C. Kaposi’s sarcoma herpesvirus/ Human herpesvirus-8 (KSHV/HHV8), and the oncogenesis of Kaposi’s sarcoma. Nat. Rev. Cancer., 2010; 10(10):707-719.
- Cesarman E and Knowles D. M. Kaposi’s sarcoma-associated herpesvirus: a lymphotropic human herpesvirus associated with Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Semin. Diagn. Pathol., 1997; 14(1): 54-66.
- Lin K, Dai C. Y and Ricciardi R. P. Cloning and functional analysis of Kaposi’s sarcoma-associated herpesvirus DNA polymerase and its processivity factor. J. Virol., 1998; 72(7): 6228-6232.
- Cunningham C, Barnard S, Blackbourn D. J and Davison A. J. Transcription mapping of human herpesvirus 8 genes encoding viral interferon regulatory factors. J. Gen. Virol., 2003; 84(6): 1471-1483.
- Habbu P, Warad V, Shastri R, Madagundi S and Kulkarni V.H. Antimicrobial metabolites from marine microorganisms. Chin. J. Nat. Med., 2016; 14(2): 101-116.
- Ponnusamy R, Petoukhov M. V, Correia B, Custodio T. F, Juillard F, Tan M, Pires de Miranda M, Carrondo M. A, Simas J. P, Kaye K. M, Svergun D and McVey C. E. KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA. Nucleic Acids Res., 2015; 43(20): 10039-10054.
- Baltz J. L, Filman D. J, Ciustea M, Silverman J. E, Lautenschlager C. L, Coen D. M, Ricciardi R. P and Hogle J. M. The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol., 2009; 83(23): 12215-12228.
- Webb B and Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinformatics., 2016; 54: 5.6.1-5.6.37.
- Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L and Schwede T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017; 45(D1): D313-D319.
- Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti-Renom MA, Davis FP and Sali A. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res., 2009; 37(Database issue): D347-54.
- O’Boyle N. M, Banck M, James C. A, Morley C, Vandermeersch T and Hutchison G. R. Open Babel: An open chemical toolbox. J. Cheminform., 2011; 3: 33.
- Khan S. A, Kumar S and Ali M. M. Virtual Screening of Molecular Properties and Bioactivity Score of Boswellic Acid Derivatives in Search of Potent Anti-Inflammatory Lead Molecule. Int. J. Interdiscip. Multidiscip. Stud., 2013; 1(1): 8-12.
- Fernandez-Recio J, Totrov M and Abagyan R. Screened charge electrostatic model in protein-protein docking simulations. Pac. Symp. Biocomput., 2002; 7: 552-563.
- Lipinski C. A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol., 2004; 1(4): 337-341.
- Aparoy P, Reddy K. K and Reddanna P. Structure and ligand based drug design strategies in the development of novel 5- LOX inhibitors. Curr. Med. Chem., 2012; 19(22): 3763-3778.
- Lovell S. C, Davis I. W, Arendall W. B 3rd, de Bakker P. I, Word J. M, Prisant M. G, Richardson J. S and Richardson D. C. Structure validation by C alpha geometry: phi, psi and C beta deviation. Proteins., 2003; 50(3): 437-450.
- Wiederstein M and Sippl M. J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res., 2007; 35 (Web Server issue):W407-W410.
- Satpathy R, Behera R and Guru R. Homology modelling and molecular dynamics study of plant defensin DM-AMP1. J. Biochem. Tech., 2012; 3(4): 309-311.
- Prajapat R, Bhattacharya I and Kumar A. Homology Modeling and Structural Validation of Type 2 Diabetes Associated Transcription Factor 7-like 2 (TCF7L2). Trends in Bioinformatics., 2016; 9: 23-29.
- Johansson M. U, Zoete V, Michielin O and Guex N. Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics., 2012; 13(1): 173.
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y and Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res., 2006; 34(Web Server issue): W116-W118.
- Narang S. S, Shuaib S and Goyal B. Molecular insights into the inhibitory mechanism of rifamycin SV against β2-microglobulin aggregation: A molecular dynamics simulation study. Int. J. Biol. Macromol., 2017; 102: 1025-1034.
- Sundarrajan S, Lulu S and Arumugam M. Computational evaluation of phytocompounds for combating drug resistant tuberculosis by multi-targeted therapy. J. Mol. Model., 2015; 21(9): 247.
- Keller S, Nicholson G, Drahl C, Sorensen E, Fiedler H. P and Süssmuth R. D. Abyssomicins G and H and atropabyssomicin C from the marine Verrucosisporastrain AB-18-032. J. Antibiot., (Tokyo) 2007; 60: 391-394.
- Kreuter M. H, Robitzki A, Chang S, Steffen R, Michaelis M, Kljajić Z, Bachmann M, Schröder H. C and Müller W. E. G. Production of the cytostatic agent aeroplysinin by the sponge Verongia aerophoba in in vitro culture. Comp. Biochem. Physiol., 1992; 101: 183-187.
- Barros F. C, da Silva D. C, Sombra V. G, Maciel J. S, Feitosa J. P, Freitas A. L and de Paula R. C. Structural characterization of polysaccharide obtained from red seaweed Gracilaria caudata (J Agardh). Carbohydr. Polym., 2013; 92: 598-603.
- Novoa-Garrido M, Aanensen L, Lind V, Larsen H. J. S, Jensen S. K, Govasmark E and Steinshamn. Immunological effects of feeding macroalgae and various vitamin e supplements in Norwegian white sheep-ewes and their offspring. Livest. Sci., 2014; 167: 126-136.
- Chlipala G. E, Pham H. T, Nguyen V. H, Krunic A, Shim S. H, Soejarto D. D and Orjala J. Nhatrangins A and B, Aplysiatoxin-Related Metabolites from the Marine Cyanobacterium Lyngbya majuscula from Vietnam. J. Nat. Prod., 2010; 73(4): 784-787.
- Rajesh R. P and Annappan M. Anticancer effects of brominated indole alkaloid Eudistomin H from marine ascidian Eudistoma viride against cervical cancer cells (HeLa). Anticancer Res., 2015; 35(1): 283-293.
- Liu X, Yuan W. Q, Sharma-Shivappa R and van Zanten J. Antioxidant activity of phlorotannins from brown algae. Int. J. Agric. & Biol. Eng., 2017; 10(6): 184-191.
- Shah M. M. R, Liang Y, Cheng J. J and Daroch M. Astaxanthin-producing green microalga Haematococcus pluvialis: from single cell to high value commercial products. Front. Plant Sci., 2016; 7: 531.
- Mitchell S. S, Nicholson B, Teisan S, Lam K. S and Potts B. C. Aureoverticillactam, a novel 22-atom macrocyclic lactam from the marine actinomycete Streptomyces aureoverticillatus. J. Nat. Prod., 2004; 67: 1400-1402.
- Xu Y, Ibrahim I. M, Wosu C. I, Ben-Amotz A and Harvey P. J. Potential of New Isolates of Dunaliella Salina for Natural β-Carotene Production. Biology., 2018; 7(1): 14.
- Bonfim-Mendonça P de S, Capoci I. R. G, Tobaldini-Valerio F. K, Negri M and Svidzinski T. I. E. Overview of β-Glucans from Laminaria spp.: Immunomodulation Properties and Applications on Biologic Models. Int. J. Mol. Sci., 2017; 18(9): 1629.
- Stritzke K, Schulz S, Laatsch H, Helmke E and Beil W. Novel caprolactones from a marine Streptomycete. J. Nat. Prod., 2004; 67: 395-401.
- Maskey R. P, Li F. C, Qin S, Fiebig H. H and Laatsch H. Chandrananimycins A ∼ C: production of novel anticancer antibiotics from a marine Actinomadura sp. isolate M048 by variation of medium composition and growth conditions. J. Antibiot., (Tokyo) 2003; 56: 622-629.
- Tsuda M, Kasai Y, Komatsu K, Sone T, Tanaka M, Mikami Y and Kobayashi J. Citrinadin, A, a novel pentacyclic alkaloid from marine‐derived fungus Penicillium citrinum. Org. Lett., 2004; 6: 3087-3089.
- Jones A. C, Monroe E. A, Podell S, Hess W. R, Klages S, Esquenazi E, Niessen S, Hoover H, Rothmann M, Lasken R. S, Yates J. R, Reinhardt R, Kube M, Burkart M. D, Allen E. E, Dorrestein P. C, Gerwick W. H and Gerwick L. Genomic insights into the physiology and ecology of the marine filamentous cyanobacterium Lyngbya majuscula. Proc. Natl. Acad. Sci., 2011; 108: 8815-8820.
- Sanchez-Machado D. I, Lopez-Hernandez J, Paseiro-Losada P and Lopez-Cervantes J. An HPLC method for the quantification of sterols in edible seaweeds. Biomed. Chromatogr., 2004; 18: 183-190.
- Warabi K, Matsunaga S, van Soest R. W. M and Fusetani N. Dictyodendrins A-E, the First Telomerase-Inhibitory Marine Natural Products from the Sponge Dictyodendrilla verongiformis. J. Org. Chem., 2003; 68(7): 2765-2770.
- Ayyad S. E. N, Makki M. S, Al-kayal N. S, Basaif S. A, El-Foty K. O, Asiri A. M, Alarif W. M and Badria F. A. Cytotoxic and protective DNA damage of three new diterpenoids from the brown alga Dictyota dichotoma. Eur. J. Med. Chem., 2011a; 46: 175-182.
- Alarado A. B and Gerwick W. H. Dictyol H, a new tricyclic diterpenoid from the brown seaweed Dictyota dentata. J. Nat. Prod., 2004; 48: 132-134.
- Cichewicz R. H, Clifford L. J, Lassen P. R, Cao X, Freedman T. B, Nafie L. A, Deschamps J. D, Kenyon V. A, Flanary J. R, Holman T. R and Crews P. Stereochemical determination and bioactivity assessment of (S)-(+)-curcuphenol dimers isolated from the marine sponge Didiscus aceratus and synthesized through laccase biocatalysis. Bioorg. Med. Chem., 2005; 13: 5600-5612.
- Ruiz C, Valderrama K, Zea S and Castellanos L. Mariculture and Natural Production of the Antitumoural (+)-Discodermolide by the Caribbean Marine Sponge Discodermia dissoluta. Mar. Biotechnol., 2013; 15(5): 571-583.
- Simmons T. L, Nogle L. M, Media J, Valeriote F. A, Mooberry S. L and Gerwick W. H. Desmethoxymajusculamide C, a cyanobacterial depsipeptide with potent cytotoxicity in both cyclic and ring-opened forms. J. Nat. Prod., 2009; 72:1011-1016.
- Hadley K. B, Bauer J and Milgram N. W. The oil-rich alga Schizochytrium sp. as a dietary source of docosahexaenoic acid improves shape discrimination learning associated with visual processing in a canine model of senescence. Prostaglandins Leukot. Essent. Fatty Acids., 2017; 118: 10-18.
- Ioannou E, Quesada A, Rahman M. M, Gibbons S, Vagias C and Roussis V. Dolabellanes with antibacterial activity from the brown alga Dilophus spiralis. J. Nat. Prod., 2011; 74: 213-222.
- Williams D. E, Patrick B. O, Behrisch H. W, Soest R. V, Roberge M and Andersen R. J. Dominicin, a cyclic octapeptide, and laughine, a bromopyrrole alkaloid, isolated from the Caribbean marine sponge Eurypon laughlini. J. Nat. Prod., 2005; 68: 327-330.
- Bai R. L, Paull K. D, Herald C. L, Malspeis L, Pettit G. R and Hamel E. Halichondrin B and homohalichondrin B, marine natural products binding in the Vinca domain of tubulin. Discovery of tubulin-based mechanism of action by analysis of differential cytotoxicity data. J. Biol. Chem., 1991; 266: 15882-15889.
- Mühlroth A, Li K, Røkke G, Winge P, Olsen Y, Hohmann-Marriott M. F, Vadstein O and Bones A. M. Pathways of Lipid Metabolism in Marine Algae, Co-Expression Network, Bottlenecks and Candidate Genes for Enhanced Production of EPA and DHA in Species of Chromista. Mar. Drugs., 2013; 11(11): 4662-4697.
- Abad J. L, Nieves I, Rayo P, Casas J, Fabriàs G and Delgado A. Straightforward access to spisulosine and 4,5-dehydrospisulosine stereoisomers: probes for profiling ceramide synthase activities in intact cells. Org. Chem., 2013; 78 (12): 5858-5866.
- Bruntner C, Binder T, Pathom-aree W, Goodfellow M, Bull A. T, Potterat O, Puder C, Hörer S, Schmid A, Bolek W, Wagner K, Mihm G and Fiedler H. P. Frigocyclonine, a novel angucyclinone antibiotic produced by a Streptomyces griseus strain from Antarctica. J. Antibiot., 2005; 58: 346-349.
- Fletcher H. R, Biller P, Ross A. B and Adams J. M. M. The seasonal variation of fucoidan within three species of brown macroalgae. Algal Res., 2017; 22: 79-86.
- Sanchez-Machado D, Lopez-Hernandez J, Paseiro-Losada P and Lopez-Cervantes J. An HPLC method for the quantification of sterols in edible seaweeds. Biomed. Chromatogr., 2004; 18: 183-190.
- Shannon E and Abu-Ghannam N. Enzymatic extraction of fucoxanthin from brown seaweeds. Int. J. Food Sci. Technol., 2018; 53(9): 2195-2204.
- Macherla V. R, Liu J, Bellows C, Teisan S, Nicholson B, Lam K. S and Potts B. C. Glaciapyrroles A, B, and C, pyrrolosesquiterpenes from a Streptomyces sp. isolated from an Alaskan marine sediment. J. Nat. Prod., 2005; 68: 780-783.
- Mori T, O’Keefe B. R, Sowder R. C, Bringans S, Gardella R, Berg S, Cochran P, Turpin J. A, Buckheit R. W Jr, McMahon J. B and Boyd M. R. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem., 2005; 280: 9345-9353.
- Fitzner A, Frauendorf H, Laatsch H and Diederichsen U. Formation of gutingimycin: analytical investigation of trioxacarcin A-mediated alkylation of dsDNA. Anal. Bioanal. Chem., 2008; 390(4): 1139-1147.
- Asolkar R. N, Schröder D, Heckmann R, Lang S, Wagner-Döbler I and Laatsch H. Helquinoline, a new tetrahydroquinoline antibiotic from Janibacter limosus Hel1. J. Antibiot., (Tokyo) 2004; 57: 17-23.
- Maskey R. P, Helmke E and Laatsch H. Himalomycin A and B: isolation and structure elucidation of new fridamycin type antibiotics from a marine Streptomyces isolate. J. Antibiot., (Tokyo) 2003; 56: 942-949.
- Loganzo F, Discafani C. M, Annable T, Beyer C, Musto S, Hari M, Tan X, Hardy C, Hernandez R, Baxter M, Singanallore T, Khafizova G, Poruchynsky M. S, Fojo T, Nieman J. A, Ayral-Kaloustian S, Zask A, Andersen R. J and Greenberger L. M. HTI-286, a synthetic analogue of the tripeptide hemiasterlin, is a potent antimicrotubule agent that circumvents P-glycoprotein-mediated resistance in vitro and in vivo. Cancer Res., 2003; 63(8): 1838-1845.
- Kusama T, Tanaka N, Takahashi-Nakaguchi A, Gonoi T, Fromont J and Kobayashi J. Bromopyrrole alkaloids from a marine sponge Agelas sp. Chem. Pharm. Bull., 2014; 62(5): 499-503.
- Itoh T, Kinoshita M, Aoki S and Kobayashi M. Komodoquinone A, a novel neuritogenic anthracycline from marine Streptomyces sp. KS3. J. Nat. Prod., 2003; 66: 1373-1377.
- García-Ruiz C and Sarabia F. Chemistry and Biology of Bengamides and Bengazoles, Bioactive Natural Products from Jaspis Sponges. Mar. Drugs., 2014; 12(3): 1580-1622.
- Manam R. R, Teisan S, White D. J, Nicholson B, Grodberg J, Neuteboom S. T. C, Lam K. S, Mosca D. A, Lloyd G. K and Potts B. C. M. Lajollamycin, a Nitro-tetraene Spiro- β-lactone-γ-lactam antibiotic from the marine actinomycete Streptomyces nodosus. J. Nat. Prod., 2005; 68(2): 240-243.
- Diogo J. V, Novo S. G, González M. J, Ciancia M and Bratanich A. C. Antiviral activity of lambda-carrageenan prepared from red seaweed (Gigartina skottsbergii) against BoHV-1 and SuHV-1. Res. Vet. Sci., 2015; 98: 142-144.
- Facompré M, Tardy C, Bal-Mahieu C, Colson P, Perez C, Manzanares I, Cuevas C and Bailly C. Lamellarin D: a novel potent inhibitor of topoisomerase I. Cancer Res., 2003; 63(21): 7392-7399.
- Kadam S. U, O’Donnell C. P, Rai D. K, Hossain M. B, Burgess C. M, Walsh D and Tiwari B. K. Laminarin from irish brown seaweeds Ascophyllum nodosum and Laminaria hyperborea: Ultrasound assisted extraction, characterization and bioactivity. Mar. Drugs., 2015; 13: 4270-4280.
- Mooberry S. L, Randall-Hlubek D. A, Leal R. M, Hegde S. G, Hubbard R. D, Zhang L and Wender P. A. Microtubule-stabilizing agents based on designed laulimalide analogues. Proc. Natl. Acad. Sci., 2004; 101(23): 8803-8808.
- Sun J, Shi D, Ma M, Li S, Wang S, Han L, Yang Y, Fan X, Shi J and He L. Sesquiterpenes from the red alga Laurencia tristicha. J. Nat. Prod., 2005b; 68(6): 915-919.
- Del Campo J. A, Rodríguez H, Moreno J, Vargas M. A, Rivas J and Guerrero M. G. Lutein production by Muriellopsis sp. in an outdoor tubular photobioreactor. J. Biotechnol., 2001; 81: 289-295.
- Kwon H. C, Kauffman C. A, Jensen P. R and Fenical W. Marinomycins A-D antitumor antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora”. J. Am. Chem. Soc., 2006; 128: 1622-1632.
- Kanoh K, Matsuo Y, Adachi K, Imagawa H, Nishizawa M and Shizuri Y. Mechercharmycins A and B cytotoxic substances from marine derived Thermoactinomyces sp. YM 3-251. J. Antibiot., (Tokyo) 2005; 58: 289-292.
- Shin J, Seo Y, Lee H. S, Rho J. R and Mo S. J. A new cyclic peptide from a marine derived bacterium of the genus Nocardiopsis. J. Nat. Prod., 2003; 66(6): 883-884.
- Kanoh K, Matsuo Y, Adachi K, Imagawa H, Nishizawa M and Shizuri Y. Neopetrosiamides, Peptides from the Marine Sponge Neopetrosia sp. That Inhibit Amoeboid Invasion by Human Tumor Cells. Org. Lett., 2005; 7(19): 4173-4176.
- Coba F. D. L, Aguilera J, Figueroa F. L, de Gálvez M. V and Herrera E. Antioxidant activity of mycosporine-like amino acids isolated from three red macroalgae and one marine lichen. J. Appl. Phycol., 2009; 21(2): 161-169.
- Kanakkanthara A, Northcote P. T and Miller J. H. Peloruside A: a lead non-taxoid-site microtubule-stabilizing agent with potential activity against cancer, neurodegeneration, and autoimmune disease. Nat. Prod. Rep., 2016; 33(4): 549-561.
- You H. N, Lee H. A, Park M. H, Lee J. H and Han J. S. Phlorofucofuroeckol A isolated from Ecklonia cava alleviates postprandial hyperglycemia in diabetic mice. Eur. J. Pharmacol., 2015; 752: 92-96.
- Kim S, Lee M. S, Lee B, Gwon W, Joung E, Yoon N and Kim H. Anti-inflammatory effects of sargachromenol-rich ethanolic extract of Myagropsis myagroides on lipopolysaccharide-stimulated BV-2 cells. BMC Complement Alternat. Med., 2014; 14: 231.
- Li Y, Fu X, Duan D, Liu X, Xu J and Gao X. Extraction and Identification of Phlorotannins from the Brown Alga, Sargassum fusiforme (Harvey) Setchell. Jacobson PB, ed. Mar. Drugs., 2017; 15(2): 49.
- Kuddus M, Singh P, Thomas G and Al-Hazimi A. Recent Developments in Production and Biotechnological Applications of C-Phycocyanin. Biomed. Res. Int., 2013; 2013: 742859.
- Glazer A. N and Hixson C. S. Subunit structure and chromophore composition of rhodophytan phycoerythrins. Porphyridium cruentum B-phycoerythrin and b-phycoerythrin. J. boil. Chem., 1977; 252(1): 32-42.
- Li J, Li C, Riccio R, Lauro G, Bifulco G, Li T. J, Tang H, Zhuang C. L, Ma H, Sun P and Zhang W. Chemistry and Selective Tumor Cell Growth Inhibitory Activity of Polyketides from the South China Sea Sponge Plakortis sp. Zhou Y-D, Nagle DG, eds. Mar. Drugs., 2017; 15(5): 129.
- Erickson K. L, Beutler J. A, Cardellina J. H and Boyd M. R. Salicylihalamides A and B, Novel Cytotoxic Macrolides from the Marine Sponge Haliclona sp. J. Org. Chem., 1997; 62(23): 8188-8192.
- Feling R. H, Buchanan G. O, Mincer T. J, Kauffman C. A, Jensen P. R and Fenical W. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed., 2003; 42(3): 355-357.
- D’Anbrosio M, Guerriero A and Pietra F. Isolation from the Mediterranean Stolonifern Coral Sarcodictyon roseum of Sarcodictyin C, D, E, and F, novel diterpenodic alcohols esterified by (E)- or (Z)-N(1)-methylurocanic acid. Failure of the carbon-skeleton type as a classification criterion. Helv. Chim. Acta., 1988; 71(5): 964-976.
- Erba E, Bergamaschi D, Ronzoni S, Faretta M, Taverna S, Bonfanti M, Catapano C. V, Faircloth G, Jimeno J and D’Incalci M. Mode of action of thiocoraline, a natural marine compound with anti-tumour activity. Br. J. Cancer., 1999; 80(7): 971-980.
- Maskey R. P, Helmke E, Kayser O, Fiebig H. H, Maier A, Busche A and Laatsch H. Anti-cancer and Antibacterial Trioxacarcins with High Anti-malaria Activity from a Marine Streptomycete and their Absolute Stereochemistry. J. Antibiot., (Tokyo) 2004; 57(12): 771-779.
- Simone M, Erba E, Damia G, Vikhanskaya F, Di Francesco A. M, Riccardi R, Bailly C, Cuevas C, Fernandez Sousa-Faro J. M and D’Incalci M. Variolin B and its derivate deoxy-variolin B: New marine natural compounds with cyclin-dependent kinase inhibitor activity. Eur. J. Cancer., 2005; 41(15): 2366-2377.
- Rajauria G and Abu-Ghannam N. Isolation and Partial Characterization of Bioactive Fucoxanthin from Himanthalia elongata Brown Seaweed: A TLC-Based Approach. Int. J. Anal. Chem., 2013; 2013: 802573.








