Raman Jasrotia* and Seema Langer
and Seema Langer
Animal Cytogenetics Lab, Department of Zoology, University of Jammu, Jammu (180006), India.
Corresponding Author E-mail: ramanjasrotia18@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1749
Abstract
Genetic variations among prawns act as an important tool to characterize and differentiate between the species. Molecular and phylogenetic analysis of shrimps and prawns like any other organism rely on high yields of pure and better quality genomic DNA. In this regard isolation of DNA is the first and basic step. In spite of the availability of many protocols of DNA extraction from animal tissues, it is difficult to ascertain that which one would provide desired results for prawn tissue. In the present study, three different techniques of DNA isolation i.e., salting out, phenol-chloroform and Qiagen DNA extraction kit were performed and compared for their yield. Cephalothoracic tissue and muscle tissue of pleopods were used for isolation. Tissue samples from fresh specimens as well as from alcohol preserved specimens were employed for extraction. The quantity (µg/ml) and quality of isolated DNA were determined by UV spectrophotometry and agarose gel electrophoresis. Results showed that Phenol-chloroform method with slight modifications obtained higher yield of genomic DNA as compared to other methods. The present work also revealed that among fresh specimens cephalotoracic tissue yielded high concentration DNA than muscle tissue. However, among alcohol preserved specimens, the concentration of DNA was higher in muscle tissue of pleopods. The high quality DNA was then subjected to randomly amplified polymorphic DNA (RAPD) and inter simple sequence repeats (ISSR) analysis. The DNAs produced clear, sharp and reproducible PCR (Polymerse chain reaction) product pattern.
Keywords
DNA Extraction; Genetic Variations; ISSR; PCR; Phenol-Chloroform; RAPD; Spectrophotometry
Download this article as:| Copy the following to cite this article: Jasrotia R, Langer S. Comparative Account of DNA Extraction Protocols in Some Fresh Water Prawns of Genus Macrobrachium (Bate, 1868) (Family Palaemonidae) from Jammu Waters for PCR Based Applications. Biomed Pharmacol J 2019;12(3). |
| Copy the following to cite this URL: Jasrotia R, Langer S. Comparative Account of DNA Extraction Protocols in Some Fresh Water Prawns of Genus Macrobrachium (Bate, 1868) (Family Palaemonidae) from Jammu Waters for PCR Based Applications. Biomed Pharmacol J 2019;12(3). Available from: https://bit.ly/2YdAAFa |
Introduction
Aquaculture has become an emerging field to meet the nutritional and economic needs of man in 21st century. In this context, culture of fishes and shell fishes (prawns, shrimps and crabs) on a large scale is contributing significantly to achieve global food security targets. The fresh water prawns besides their high dietary value have significant medical importance too as some of the Macrobrachium species serve an imperative role in the biological control of human schistosomiasis by acting as predators of the snail species which are the intermediate hosts of the parasite Schistosoma.1,2
In Jammu division of J&K state, several prawn species such as Macrobrachium dayanum, M. kistensis and M. lamarrei are on record3 whose nutritional value is at par with other culturable fish species.4 Prawn and shrimp farming requires suitable candidates which can withstand captive conditions, has higher genetic diversity and genetic variability to adapt to different environments. In this regard the extent of variability needs to be screened through RAPD and ISSR studies and thus extraction of Genomic DNA is prerequisite for any DNA based investigation.5
The protocol for DNA extraction must be simple, inexpensive, reliable, quick and safe with minimal risk for the user.6,7 For PCR amplification quality of the Genomic DNA is crucial as excess of cell debris and proteins may inhibit the amplification process.6 That’s why efficient DNA isolation methods have been a core element of molecular research. In the current study, three protocols such as organic, inorganic and kit method were evaluated and compared for total DNA isolation from Macrobrachium sp of Jammu waters.
Material and Methods
Specimens of prawns were collected from Sehi stream (32° 30’ N, 74° 43’ E) of Jammu district and Kheri stream of Samba district (32° 37’ N, 74° 52’ E) by using cast net of mesh size 5mm x 5mm and brought to Animal Cytogenetics lab of Department of Zoology, University of Jammu. The live specimens were immediately used for DNA isolation and dead specimens were preserved in 75 % ethyl alcohol.
50 mg of cephalothoracic tissue and 50 mg of muscle tissue of pleopods from fresh as well as alcohol preserved specimens were used for extraction of total DNA. Before homogenizing the tissue, ethyl alcohol was removed to prevent destabilization of the isolated DNA. Three methods of DNA isolation were used and compared for their yield.
Protocol 1
Total DNA was extracted using Salting out method with slight modifications.9 50 mg of tissue from each sample was homogenized using mortar and pestle. It was then transferred to microcentrifuge tubes with cell lysis solution containing 10 mM Tris-HCl (pH 8.0), 100 mM EDTA (pH 8.0), 2% SDS (Sodium dodecyl sulphate) with pH 8.0, 0.5 M NaCl. Then 5 µl of proteinase K (20 mg/ml) was added in each tube. The samples were incubated at 60°C for 10-12 hours (with periodic mixing). After that 6M NaCl (saturated NaCl) was added to each tube and subjected to centrifugation at 8000 rpm for 10 minutes. The supernatant was collected and transferred to other microcentrifuge tube. DNA was then precipitated using absolute alcohol. The DNA pellet was dissolved in 200 µl TE buffer (10 mM Tris-HCl, 1 mM EDTA).
Protocol 2
Genomic DNA was isolated using standard Phenol-Chloroform method with minor changes.10 After homogenization of 50 mg of tissue, each sample was exposed to the treatment of lysis buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM EDTA (pH 8.0), 100 mM NaCl, 1 % SDS (pH 8.0), 5M NaCl. Samples were then incubated with proteinase K (20 mg/ml) at 55°C overnight and the lysate was centrifuged at 10,000 rpm for 10 minutes and washed with phenol: chloroform: isoamyl alcohol (25:24:1). In the supernatant the DNA was precipitated with chilled isopropanol and mixed by inversion. DNA pellet was then washed with 70 % ethanol and air dried. After that the extracted DNA was dissolved in 200 µl TE buffer (10 mM Tris-HCl, 1 mM EDTA).
Protocol 3
The DNA was extracted using kit method (DNeasy Blood & Tissue Kit, Qiagen, Germany) following the Kit- manufacturer’s instructions with modifications. To about 25-50 mg of tissue from each sample in a 1.5 ml microcentrifuge tube, Buffer ATL (Lysis buffer) was added along with 20 µl of Proteinase K and incubated at 56°C for 10-15 minutes until completely lysed. Then after few treatments with other buffer solutions, the DNA was finally eluted on a spin column membrane.
Evaluation of DNA Purity, Quality and Quantity
Agarose gel electrophoresis is a standard method to determine the quality of Genomic DNA as it separates and recognizes DNA fragments according to their molecular weights. 1 % Agarose gel was prepared to check the total DNA of prawns. Clear and sharp bands near the wells indicated high molecular weight DNA. The agarose gel was then photographed with high resolution camera. The quantity of Genomic DNAs isolated by three protocols were determined by taking the absorbance reading at wavelength of 260 nm on UV spectrophotometer and the purity of DNA was analysed by calculating the ratio of sample absorbance at 260 and 280 nm (260/280).11,12,13 The DNA concentration (C) was determined following the formula: Concentration (C) = A260 × 50 µgml-1 × dilution factor.12,14,15 A 50 µg ml-1 solution of double stranded DNA gives the optical density reading of 1.0 at 260 nm.11
Amplification by PCR
The Genomic DNA isolated by three protcols were subjected to PCR amplification by RAPD (5′-CAGGCCCTTC-3′) and ISSR (5′-CACACACACACACACAAT-3′) primers. The polymerase chain reaction was performed using 2 μl DNA, 2.5 μl reaction buffer, 1.0 μl dNTPs (10 mM), 1.0 μl Taq Polymerase (1U/μl), 2.5 μl MgCl2 (25 mM), 2.0 μl primer and 14 μl PCR water to make up the final rxn volume of 25 μl. The conditions used for the amplification were as follows: an initial denaturation step at 95°C for 5 minutes; followed by 45 cycles of 94°C for 1 minute, 50°C for 1 minute and 72°C for 1 minute; and finally elongation (extension) at 72°C for 10 minutes.
The results were presented as means ±SD and were processed using Statistical Package for Social Sciences (SPSS) software version 20.
Results and Discussion
Among the three studied species of prawns (Fig. 1a, 1b, 1c) Macrobrachium dayanum yielded high quality DNA in all the extraction protocols. DNA concentration, however, in two species viz., M. dayanum and M. kistensis was observed to be highest following the Phenol-Chloroform method as compared to salting out and kit method where as in third species M. lamarrei high concentration DNA was extracted by kit method as depicted in Table 1. A concentration range between 180 to 315 µg/ml, 67.5 to 752.5 µg/ml and 42.5 to 365 µg/ml was found using salting out, phenol-chloroform and kit methods respectively, thereby revealing phenol-chloroform method to be the best for DNA extraction of fresh water prawns. The purity of Genomic DNA isolated by three protocols was determined by ratio of optical density readings at 260 nm and 280 nm on UV spectrophotometer as shown in Table 2.
The high quality DNA isolated from above methods was subjected to PCR amplification by RAPD and ISSR markers. The electrophoretic pattern of genomic DNA showed single sharp and distinct band on 1 % agarose gel for each sample and the electrophoretic images of PCR products showed many sharp and distinct bands on 1.8 and 2 % agarose gel for prawn populations as depicted in figure 2 and figure 3.
 |
Figure 1a: Macrobrachium dayanum. 1b: Macrobrachium kistensis. 1c: Macrobrachium lamarrei.
|
 |
Figure 2: DNA isolated by three methods; 2a: Salting out, 2b: Phenol-Chloroform method, 2c: Kit method (Lane M represents DNA ladder, Lane 1,2 and 3 depicts DNA of three species of Macrobrachium).
|
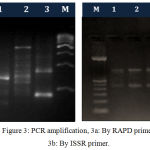 |
Figure 3: PCR amplification, 3a: By RAPD primer, 3b: By ISSR primer.
|
Table 1: Mean values of DNA concentration by three protocols.
| Prawn species | DNA Concentration (µg/ml) | ||
| Salting out method | Phenol-Chloroform method | Kit method | |
| Macrobrachium dayanum | 315±0.77 | 752.5±0.68 | 42.5±0.8 |
| Macrobrachium lamarrei | 180±1.2 | 67.5±0.90 | 335±1.0 |
| Macrobrachium kistensis | 215±0.78 | 540±1.1 | 365±0.88 |
Table 2: Mean values of optical density for estimation of purity of DNA.
| Prawn specimens | DNA extracted by Salting out | DNA extracted by Phenol-Chloroform | DNA extracted by Qiagen kit | |||
| OD260 | OD260/280 | OD260 | OD260/280 | OD260 | OD260/280 | |
| Macrobrachium dayanum | 0.126±0.0005 | 1.82±0.0008 | 0.301±0.0006 | 1.79±0.0005 | 0.017±0.0009 | 1.88±0.0005 |
| Macrobrachium lamarrei | 0.072±0.0007 | 1.74±0.0009 | 0.027±0.0008 | 1.86±0.0009 | 0.134±0.0005 | 1.83±0.0002 |
| Macrobrachium kistensis | 0.860±0.0005 | 1.83±0.0009 | 0.216±0.0005 | 1.89±0.0008 | 0.146±0.0002 | 1.71±0.0005 |
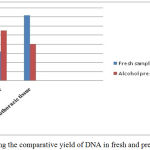 |
Graph 1: Showing the comparative yield of DNA in fresh and preserved samples.
|
The two different tissues viz., cephaothoracic tissue after removal of carapace and muscle tissue of pleopods were used for isolation of DNA. The quantity of DNA in cephalothoracic tissue was higher compared to muscle tissue in the freshly collected prawn samples whereas the DNA concentration was higher in muscle tissue than the other tissue as shown in the graph 1. The reason for this can be attributed to the fact that the hepatopancreas in the cephaothoracic region is light orange in colour. It is known to possess numerous chromatins which are supposed to contribute in the absorbance spectrum thereby increasing yield.5 Regarding the purity and quality of DNA the value of OD 260/280nm around 1.8 is considered best.16,17,18 The value of OD 260/280nm less than 1.8 specify protein or phenol contamination and value higher than 1.8 indicate RNA contamination.5,15 The phenol-chloroform method has also been found high yielding for DNA isolation in shrimps as compared to other two methods.5,19 Also, this method is used commonly for extraction of gDNA in studies pertaining to taxonomy and detection of white spot syndrome viruses in several species of prawns and shrimps as well as other crustaceans.20,21,22
Conclusion
The present study depicts that Phenol-Chloroform method is best to isolate DNA from fresh water prawns. The genomic DNA obtained by it provided better PCR results for RAPD and ISSR analysis of genetic diversity of prawns.
Acknowledgements
Authors are highly thankful to Head, Department of Zoology, University of Jammu for providing necessary lab facilities.
Conflict of Interest
There is no conflict of interest.
References
- Sokolow, S.H., Lafferty, K.D and Kuris, A.M. Regulation of laboratory populations of snails (Biomphalariaand Bulinus ) by river prawns, Macrobrachium spp. (Decapoda, Palaemonidae): implications for control of schistosomiasis. Acta Tropica, 132: 64-74 (2014).
- Sokolow, S.H., Huttinger, E., Jouanard, N., Hsieh, M.H., Lafferty, K.D., Kuris, A.M., Riveau, G., Senghor, S., Thiam, C., N’Diaye, A., Faye, S.D and De Leo, G.A. Reduced transmission of human schistosomiasis after restoration of a native river prawn that preys on the snail intermediate host. Proceedings of the National Academy of Sciences of the United States of America, 112(31): 9650–9655 (2015).
- Sharma, N., Langer, S and Jayachandran K.V. On the report of two species of genus Macrobrachium from Jammu waters. International Journal of Zoology Studies, 3(1): 91-95 (2018).
- Jasrotia, R., Langer, S., Palaq., Sharma, N and Panjaliya, R.K. Assessment of Genetic variability of two fresh water prawns, Macrobrachium dayanum and Macrobrachium lamarrei from Jammu region by using ISSR markers. International Journal of Recent Scientific Research, 8(10): 21176-21180 (2017).
- Biswas, B., Islam, M.N., Siddiqui, M.N. and Ahsan, M.N. 2008. Optimization of DNA Extraction Protocols from Shrimp (Penaeus monodon) Tissue. Khulna University Studies, 9(2): 239-242 (2008).
- Guillemaut, P., Marechal-Drouard, L. Isolation of plant DNA: a fast, inexpensive and reliable method. Plant Mol. Biol. Reptr., 10: 60-65 (1992).
- Marechal-Drouard, L., Guillemaut,P. A powerful but simple technique to prepare polysaccharide- free DNA quickly and without phenol extraction. Plant Mol. Biol. Reptr., 13: 26-30 (1995).
- Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G and Erlich, H. 1992. Specific enzymatic amplification of DNA in vitro, the polymerase chain reaction. Biotechnology, 24: 17-27 (1992).
- Aljanabi, S.M. and Martinez, I. Universal and rapid salt extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Research, 25(22): 4692-4693 (1997).
- Sambrook, J., Russell, D.W. Molecular cloning. A laboratory manual. 3rd edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York (2001).
- Sambrook, J., Fritschi, E.F and Maniatis, T. Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, New York (1989).
- Aquadro, C.F., Noon, W.A., Begun, D.J and Danforth, B.N. RFLP analysis using heterologous probes. In: Hoelzel, A.R. (ed.) Molecular Genetics Analysis of Populations. A Practical Approach. New York, Oxford University Press, p. 154 (1998).
- Gavarane, I., Kokina, I., Aksjuta, K., Barsevskis, A and Valainis, U. Optimization of DNA extraction protocol for DNA isolation from air-dried collection material for further phylogenetic analysis (Coleoptera: Carabidae). Acta Biol. Univ. Daugavp., 11(2): 141-147 (2011).
- Linacero, R., Rueda, J and Vazquez, A.M. Quantifying of DNA. Karp, A., Issac P.G and Ingram, D.S. (Eds.) Molecular Tools for Screening Biodiversity: Plants and Animals. Chapman and Hall. London, Weinheim, New York, Tokyo, Melbourne, Madras, 18-21 (1998).
- DNA extraction, purification and characterization. [URL: http://www.waksmanfoundation.org/labs/wisteb/extract.htm.] (2005).
- Hillis, D.A., Larson, A., Davis, S.K and Zimmer, E.A. Nucleic acids III: sequencing. In: Hillis, D.A. and Moritz, C. (eds.), Molecular Systematic. Sunderland, Mass, Sinauer Associates, pp. 318-370 (1990).
- Chomczynski, P., Mackey, K., Drews, R and Wilfinger, W. DNAzol: A reagent for the rapid isolation of genomic DNA. BioTechniques, 22: 550-553 (1997).
- Ligozzi, M and Fontana, R. Isolation of total DNA from bacteria and yeast. African Journal of Biotechnology, 2: 251-253 (2003).
- Besbes, N and Sadok, S. Comparison of DNA-extraction Methods Suitable for PCR- based Applications to Identify Shrimp species in Commercial products. Journal of Fisheries Sciences.com, 11(4):025-31 (2017).
- Jayabarath, J., Uma Maheshwari, R., Maanusha, J and Bhavanisowndharya, B. Detection of WSS Virus on Banana Prawn, Peneaus mergueinsis By PCR Technique. Journal of Chemical and Pharmaceutical Sciences, 10(4): 1636-1638 (2017).
- Kundu, S., Rath, S., Tyagi, K., Chakraborty, R and Kumar, V. Identification of penaeid shrimp from Chilika Lake through DNA barcoding. Mitochondrial DNA Part B: Resources, 3(1):161–165 (2018).
- Varela, E.S., Goncalves, E.D., Santos, M.D., Sampaio, I and Schneider, H. Isolation and characterization of 11 microsatellite loci in the mangrove crab, Ucidescordatus (Ocypodidae: Ucidinae). Molecular Ecology Resources, 9(5): 1395-1397 (2009).








