Manuscript accepted on :21-Nov-2018
Published online on: 03-12-2018
Plagiarism Check: Yes
Reviewed by: Gyanendra Singh
Second Review by: Dr. Arun Karnwal
Final Approval by: Dr. Ayush Dogra
Mohamed Abd ElSalam1 , Doaa Gamal2
, Doaa Gamal2 , Manal El-Said2
, Manal El-Said2 , Dalia Salem2
, Dalia Salem2 , Aisha Abu Aitta2
, Aisha Abu Aitta2  and Mamdouh S. El-Gamal1
and Mamdouh S. El-Gamal1
1Departmentof Botany and Microbiology, Faculty of Science (Boys), Al-Azhar University, Cairo, Egypt.
2Department of Microbiology, Theodor Bilharz Research Institute, Giza, Egypt.
Corresponding Author E-mail: manalmicrobiology@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1565
Abstract
Resistance to quinolone has increased significantly and one of the most reasons is plasmid-mediated quinolone resistance (PMQR). The aim of this study is to detect the prevalence of PMQR in multidrug-resistant (MDR) Gram negative bacilli and to characterize these resistance genes. A total of 420 Gram negative bacilli clinical isolates were collected from patients attending Misr children hospital. Isolates were identified by biochemical reactions, while antimicrobial susceptibility testingwas done by Kirby-Bauer disk diffusion method. Minimum inhibitory concentrations (MIC) of ciprofloxacin were detected by E-test, whereas combined test method was used to detect extended-spectrum β-lactamase (ESBL) production. QnrA, qnrB, and qnrS genes were determined by multiplex polymerase chain reaction (PCR). MDRGram negative bacilli represented 68% (268/420); most of them were recovered from blood culture specimens (21%).Among these MDR isolates21%(60/268) were ciprofloxacin resistant; with MICs >32µg/ml in 95% of the isolates.ESBL production was detected in 11.7% of the studied isolates. The qnr genes were detected in 60%. QnrS and qnrB were the detected genes in 77.8% and 16.7% of the isolates respectively. Both qnrB and qnrS genes were determined simultaneously in 5.5%.QnrB gene was found alone in only one isolate (14.3%) that was ESBL-producer. The most MDR isolates were recovered from blood culture; this confirms the occurrence of these superbugs and their ability to cause life threatening infections. The prevalence of quinolone resistant Gram negative bacilli clinical isolates is high. The mostly prevalent PMQR gene is qnrS followed by qnrB.
Keywords
ESBLs; Gram Negative Bacilli; Multiplex PCR; Plasmid-Mediated Quinolone Resistance
Download this article as:| Copy the following to cite this article: ElSalam M. A, Gamal D, El-Said M, Salem D, Aitta A. A, El-Gamal M. S. Prevalence of Plasmid-Mediated Quinolone Resistance in Multidrug-Resistant Gram Negative Bacilli in Egypt. Biomed Pharmacol J 2018;11(4). |
| Copy the following to cite this URL: ElSalam M. A, Gamal D, El-Said M, Salem D, Aitta A. A, El-Gamal M. S. Prevalence of Plasmid-Mediated Quinolone Resistance in Multidrug-Resistant Gram Negative Bacilli in Egypt. Biomed Pharmacol J 2018;11(4). Available from: http://biomedpharmajournal.org/?p=24479 |
Introduction
The incidence of serious infections due to multi drug drug-resistant (MDR) Gram-negative bacteria has increased and these infections now constitute a serious threat worldwide.1 These MDR bacteria are major causes of various types of infections; respiratory tract, urinary tract, bloodstream, and wound infections.2 These pathogens have the ability to produce various β-lactamases such as extended-spectrum β-lactamases (ESBLs).3 MDR Gram-negative bacteria worldwide become a problem for clinicians and infection control staff due to few therapeutics possibilities.4
Quinolone are a group of broad-spectrum antibiotics that are widely used in routine clinical practice.5 Low side effects, large range of activities and adequate oral absorption, makequinolone the first-line of drug options to treat many infections.6 The widespread and inappropriate use of quinolone leads to a significant increase of resistant Gram-negative isolates.7-9
Resistance to quinolone is often due to several mechanisms such as point mutations in chromosomal genes such as DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE),10 decreased permeability of efflux pumps.9,11 Recently, plasmid-mediated quinolone resistance (PMQR) has also been demonstrated in several studies worldwide.9 Three mechanisms of PMQR have been described (i) target alteration by Qnr, (ii) drug modification by the aminoglycoside acetyltransferase AAC(6′)-Ib-cr, which can reduce ciprofloxacin activity, and (iii) efflux pump activation by two quinolone efflux pumps.11-14 Qnr proteins protect DNA gyrase and topoisomerase IV from the inhibitory activity of quinolone. Currently, there are six different qnr genes: qnrA, qnrB, qnrC, qnrD,qnrS, and the most recently reported;qnrVC.13
The aim of this study is to detect the prevalence of PMQR in multidrug-resistant Gram negative bacilliisolates and to determine their resistance genes.
Materials and Methods
Clinical Bacterial Isolates
A total number of 420 Gram negative bacilli clinical isolates were collected from patients attending Misr children hospital during the period from June 2017 to February 2018. Two hundred and eighty six isolates (68%; 286/420) were multidrug resistant isolates; MDR bacteria were defined as resistant to one or more antimicrobials on three or more antimicrobial classes.15 Sixty isolates (21%; 60/268) were ciprofloxacin resistance by Kirby-Bauer disk diffusion method; they were isolated from different clinical infections as urinary tract, surgical wound, chest, blood stream, central line, and umbilical catheter infections.The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki (6th revision, 2008) as reflected in a prior approval by TBRI institutional review board (FWA 00010609).
Identification of clinical isolates
Identification of the Gram negative isolates was done by; cultural characters and biochemical reactions using biochemical test media(Triple sugar iron (TSI), Lysine iron agar (LIA), Motility indole ornithine (MIO), citrate and urease)(Oxoid, England). Storage of bacterial isolates was done by adding 0.85 ml of an overnight incubated bacterial culture of the identified Gram negative isolates in nutrient broth (Oxoid, England) to 0.15 ml sterile glycerol in sterile cryotubes, which were vortexed and stored at – 20°C.16
Antimicrobial Susceptibility Testing (AST)
Gram negative clinical isolates were screened by Kirby-Bauer (KB) disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidelines.17 Resistance to quinolones was assessed using;nalidixic acid (NA; 30 µg), ciprofloxacin (CIP; 5 µg), and levofloxacin (LEV; 5 µg). Sensitivity to other antibiotics was tested using; amikacin (AK; 30 µg), gentamicin (CN; 10 µg), ceftriaxone (CRO; 30 µg), ceftazidime (CAZ; 30 µg), cefotaxime (CTX; 30 µg) cefoperazone (CEP; 75µg), amoxicillin/clavulanic (AMC; 30µg), ampicillin/sulbactam (SAM; 20 µg), and imipenem (IPM; 10 µg) (Mast Diagnostics, U.K.).The interpretation of antimicrobial susceptibility results was according to current CLSI guidelines.17
Minimum Inhibitory Concentration (MIC) of Ciprofloxacin by E-test
E-test (AB Bio Disk Solna, Sweden) was used for detection of MICof Gram negative clinical isolates to ciprofloxacin and was performed according to the instructions ofmanufacturer. MIC interpretive criteria for ciprofloxacin CLSI17 were used to interpret results; susceptible ≤1 mg/ml, intermediate = 2 mg/ml and resistant ≥ 4 µg/ml.
Detection of Extended Spectrum β-Lactamases (ESBLs)
Screening for ESBLby disk diffusion method
The multi drug resistant Gram negative clinical isolates were screened by the KB disk diffusion method where isolates with decreased susceptibilities to cefotaxime (zone diameter of <27 mm) and/or ceftazidime (zone diameter of <22 mm) were considered probable ESBL-producing pathogens according to CLSI guidelines.17
Confirmatory Test by Combination Disk Method
Phenotypic confirmation of potential ESBL-producing isolates was performed usingcefotaxime (CTX 30 μg) and cefotaxime/ clavulanic acid disks (CTC 30/10 μg) (Bio-Rad, France) according to CLSI recommendations.17 Interpretation of antimicrobial susceptibility results was according to CLSI guidelines;if the zone size increased in the presence of the inhibitor compared with the cephalosporin alone by 5 mm or more this is indicative of an ESBL-producing strain.17 E. coli ATCC 25922 was used as negative control in this method.
Multiplex PCR for detection of qnrA, qnrB, and qnrS genes
Primers Screening and Selection by Using BLAST Algorithm[18]
Several sets of PCR primer pairs sequences corresponding to different qnrA,qnrB,andqnrS gene clusters were taken from literature19-24 and searched for homology to all qnrA, qnrB, and qnrS sequences available in Gen bank database using BLAST algorithm (www.ncbi.nlm.nih.gov). Primers with 100% homology to all retrieved qnrA, qnrB, and qnrS with no match to human genes have been selected to be synthesized (Table 1)..Extraction of bacterial DNA was performed by boiling method.20 Briefly, a bacterial colony was suspended in 200 μL of sterile distilled water and heated to 95°C for 10 min in water bath. Then, the suspensions were centrifuged at 6000 rpm for 20 minute. Supernatants were separated in sterile eppendorf tube andstored at -20°C for PCR assays. PCR reactions for all primer pairs have been carried out in multiplex using negative controls with no DNA sample. DNA extract of each isolate was added to each PCR tube to reach 50μl. PCR master mix that contained; dream Taq DNA polymerase, 2 x Dream Taq green buffer, Nucleotides mix (0.4 mM each), and 4 mM MgCl2. Amplification was carried out with the following thermal cycling (Biometra, UK) conditions; initial denaturation at 95°C for 5 min, followed by 25 cycles at 95°C for 105 sec, annealing at 56°C for 15 sec, and extension at 72°C for 15 sec.25 DNA fragments were analysed by electrophoresis in a 2% agarose gel containing 0.05 mg ethidium bromide. The gel was photographed using the gel documentation system (Cleaver, UK).
Table 1: Primers for detection PMQR genes.
| Primer name | Sequence | M.W | Ref. | ||||
| qnrA | F: 5′-AGAGGATTTCTCACGCCAGG-3′
R: 5′-TGCCAGGCACAGATCTTGAC-3′ |
580bp | 20 | ||||
| qnrB | F: 5′-GGMATHGAAATTCGCCACTG-3′
R: 5′-TTYGCBGYYCGCCAGTCGAA-3′ |
264bp | |||||
| qnrS | F: 5′-GCAAGTTCATTGAACAGGGT-3′
R: 5′-TCTAAACCGTCGAGTTCGGCG-3′ |
428bp | |||||
M.W = molecular weight, f= forward, r= reverse
Statistical Methods
The data were statistically articulated inexpressions of frequencies (number of cases) and relative frequencies (percentages). P value(probability value) less than 0.05 was considered statistically significant. All statistical estimations were performed using Microsoft Excel 2013 program (Microsoft Corporation, NY., USA) and SPSS (Statistical Package for the Social Science; IBM SPSS statistic) version 20 for Microsoft Windows.
Results
The prevalence of multidrug resistant Gram negative bacilli were 68% (286/420), among them 21% isolates (60/286) were ciprofloxacin resistant. All the 60 clinical isolates were members of Enterobacteriaceae family; they were recovered from 39 males (65%) and 21 females (35%).
Most of clinical isolates were isolated from blood culture specimens46.7% (28/60). The other isolates were taken from endotracheal tube (26.7%; 16/60), urine (13.3%; 8/60), wound swabs (6.7%; 4/60), central lines (5%; 3/60), and an umbilical catheter (1.6%; 1/60).
Identification of Clinical Isolates
Klebsiellapneumoniae (K. pneumoniae) was the most frequently isolated species (66.7%; 40/60), followed by Escherichia coli (E. coli) with isolation rate of 21.7% (13/60), then Enterobacter cloacae (E. cloacae) in11.6%(7/60).The distribution of studied isolates according to the speciesand specimen type is shown in table 2.
Table 2: Distribution of bacterial isolates according to species and specimen type.
| Species (no. of Isolates) | No. (%) of isolates according to specimens | ||||||||||
| Blood culture | Urine | Endotracheal tube | Central line | Umbilical catheter | Wound swab | ||||||
| K. pneumonia (40) | 17 (42.5) | 2 (5) | 15 (37.5) | 3 (7.5) | 1 (2.5) | 2 (5) | |||||
| E. coli (13) | 6 (46.1) | 5 (38.5) | 0(0) | 0(0) | 0(0) | 2 (15.4) | |||||
| E. cloacae (7) | 5 (71.4) | 1 (14.3) | 1 (14.3) | 0(0) | 0(0) | 0(0) | |||||
Data are expressed as numbers (N) and percent (%).
Antimicrobial Susceptibility Testing (AST)
Antibiotic susceptibility pattern of 60 multidrug resistant Enterobacteriaceae isolates to different antibiotics is shown in table 3.
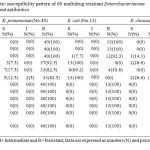 |
Table 3: Antibiotic susceptibility pattern of 60 multidrug resistant Enterobacteriaceae isolates to different antibiotics. |
Detection of ESBLs
Among the 60 clinical isolates 11.7% (7/60) isolates were confirmed phenotypically as ESBL-producers. They were E. coli isolates in 71.4%(5/60) and 2 isolates; one K. pneumoniae and another E. cloacae (14.3%, each).
E-Test
The overall prevalence of the 60 Gram negative isolates which demonstrated resistance to ciprofloxacin (MICs >32µg/ml) was 95%. They were 38 K. pneumoniae isolates (66.7%), 12 E. coli isolates (21%), and 7 E. cloacae isolates (12.3%).
The remaining isolates demonstrate different levels of resistant to ciprofloxacin; where one K. pneumoniae isolate (1.7%) was resistant to 12µg/ml and 8µg/ml respectively. One E. coli isolate (1.7%) showed intermediate susceptibility with MIC 3µg/ml.
Molecular detection of PMQR Genes
Blast alignment of selected primers: the sequences of selected primers which described previously by Cattoir et al20 were subjected to Blast alignment against the Gene Bank no redundant genes database.18 Data showed complete sequence homology and conservation of these primers with qnrA, qnrB and qnrS genes in all of the affiliated targeted species but not with non-targeted species.
Multiplex PCR detection of qnrA, qnrB, and qnrS genes in isolated bacteria
A multiplex PCR reaction was used for simultaneous detection of qnrA, qnrB and qnrS genes in each isolated bacteria (Figure 1).
The qnr genes were detected in 60% (36/60) of quinolone resistant Gram negative bacilli isolates. The qnrS was the main gene detected in 77.8% (28/36) while qnrB was detected in 16.7% (6/36). Both qnrB and qnrS genes were determined simultaneously in two K. pneumoniaeisolates (5.5%; 2/36).However, qnrA was not detected in any of the studied isolates. Quinolone resistance genes and their distribution among studied isolates according to specimens and species of the isolates are illustrated in table 4. The qnrB gene was detected in one ESBL-producer K. pneumoniae isolates (1/7; 14.3%), while qnrS gene was not detected in ESBL-producer isolate.
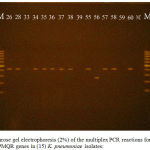 |
Figure 1: Agarose gel electrophoresis (2%) of the multiplex PCR reactions for detection of PMQR genes in (15) K. pneumoniae isolates: |
Lanes : 26,28,34,35,36,37,38,55,57,58,and 60;isolates with positive results to qnrS (427bp), lane 56;isolate with positive result to qnrB (263bp). Lanes: 33, 39, and 59 isolates show negative PCR to PMQR tested genes. Lane NC (negative control) and lanes: M; molecular size marker (Gene ruler TM 100 bp DNA ladder).
The qnr genes were detected in 60% (36/60) of quinolone resistant Gram negative bacilli isolates. The qnrS was the main gene detected in 77.8% (28/36) while qnrB was detected in 16.7% (6/36). Both qnrB and qnrS genes were determined simultaneously in two K. pneumoniaeisolates (5.5%; 2/36).However, qnrA was not detected in any of the studied isolates. Quinolone resistance genes and their distribution among studied isolates according to specimens and species of the isolates are illustrated in table 4. The qnrB gene was detected in one ESBL-producer K. pneumoniae isolates (1/7; 14.3%), while qnrS gene was not detected in ESBL-producer isolate.
 |
Table 4: Quinolone resistance genes and their distribution among studied isolates according to specimens and species of the isolate. |
Discussion
The increased prevalence of infections caused by MDR Gram-negative bacteria constitutes a serious threat to global public health due to the limited currentavailable treatment options and the lackin development of new antimicrobial agents.1 Infections caused by these pathogens are allied with elevated morbidity and mortality rates and prolonged hospital stay.2
Quinolonesis a widely used antibiotic with a broad spectrum. Inhibiting the normal functions of bacterial type II topoisomerase is the target of quinolones.10 Quinolone resistance is mainly caused by mutations in chromosomal genes encoding the quinolone targets, such as DNA gyrase and topoisomerase IV; howeverrecently PMQR geneshave been increasingly reported in most area of the world.11
The aim of this study was to determine the prevalence of PMQR among the multidrug Gram negative bacilli isolates through detection of qnr genes.
The prevalence of MDR Gram negative bacilli isolates among other similar isolates was 68% (286/420).This finding is in agreement with another study conducted in Egypt by Tohamy et al26 who identified MDR in 68.6% of isolated bacteria. This could be attributed to the irrational use of antibiotics against pathogenic bacteria.15
In this study, 60 (21%) of multidrug resistant Gram negative isolates were ciprofloxacin resistant. Most of isolates were isolated from blood culture specimens (47%). Whereas in the National Cancer Institute (NCI) in Egypt Helmy and Kashef27 reported that majority of his MDR isolates were collected from urine (36%). Our finding confirms the ability of these superbugs MDR bacteria to cause life threatening infections.
In the present study,among the 60 ciprofloxacin resistant Gram negative isolates, K. pneumoniae was the most frequently isolated species (66.7%), followed by E. coli (21.7%), then E. cloacae (12 %). These results are in accordance with Jlili et al[28] who reported that K. pneumoniae is the main species (59%) followed by E. coli (23.4%), then E. cloacae (7.9%). However, another study intertiary care cancer hospital in Cairo, Egyptstated that the most frequent isolates in immunocompromised cancer patients were ofE. coli (39.7%) followed by K. pneumoniae (34.3%%), then E. cloacae (3.3%).29 Many studies [30, 31], showedK. pneumoniaeas the most frequently Gram negative species isolated from blood cultures and this explained the majority of the K. pneumoniae isolates in this study.
Among the 60 studied clinical isolates 7 (11.7%) were ESBL-producer by combined diskmethod. These results are in accordance with Charfi et al32 who reported that, out of the 283 Gram-negative isolates, 46 (16.25%) isolates were ESBL producers, but was slightly lower than the findings ofBouchakour et al33 who reported that, out of 188 gram-negative isolates included in their study, 39 (20%) were ESBL producers. However, this figure may be underestimated, as most of our isolates (60%) were imipenem resistantand phenotypic detection of ESBL is not reliable in presence of other β-lactamases as production of carbapenemases, although not investigated in the current study, is the main mechanism of carbapenem resistance.34
In this study, qnr genes were detected in 60% (36/60) of ciprofloxacin resistant Gram negative bacilli isolates. That was consistent with another Egyptian study by Tohamy et al26 who found qnr genes in 42.9% (30/70) of MDR isolates, while that was higher than in two studies from Iran and Italy that reported a detection rate of 19% and 17% respectively.11,35
In the current study, qnr genes were found more frequently in K. pneumoniae isolates. As they were detected in 82.5% (33/40) of K. pneumoniae isolates. Nearly similar findings were reported from Italy (68.1%), Japan (66.7%), China (65.5%) and Korea (63.1%).35-38 Lower rate (52.2%) was reported in Iran.11 However, in a study conducted in both Norway and Sweden very low rate (8.3%) was reported.39 In the present study, the prevalence of qnr genes was much lower in E. coli isolates (7.7%) than in other species of Gram negative bacilli isolates. These results are in accordance with the findings of many studies that reported a low detection rate of qnr genes in E. coli isolates.11,24,35
In this study, qnrS was the main qnr gene (77.8%) found. This was in accordance to Poirel et al40 who identified qnrS in 61.5% of the Enterobacteriaceae isolates. The qnrS was detected in the current study in 62.5% of K. pneumoniaeandin 7.7% of E.coliisolates. Lower prevalence rate was determined in a study conducted in Egypt in whichqnrSwas found in 9.5% of K. pneumoniaeisolates, and in 2.7% of E.coli isolates.27 While a higher rate of qnrS(16.6%) was detected in ESBL producing E. coli isolates in another Egyptian study.41
In our study, the qnrS was detected in 28.6% of E. cloacae. This was in accordance with Yang et al37 who reported its presence in 17.1% of E. cloacae isolates. However, other studies in Korea and Morocco reported absence of qnrS in E. cloacae isolates.33,42
In the current study, qnrB was the second predominant qnrdetected gene (16.7%); qnrB gene alone was only detected in 10% of K. pneumoniae. Similarly, a study from Spain identified qnrB gene in 14.3 % of the Gram negative bacilli isolates.21 An Egyptian study reported a detection of qnrB gene in 9.5% of K. pneumoniaeisolates.27 Higher values were reported from Morocco in whichqnrBgene was found in 28.5% of K. pneumoniaeisolates.33 It worth to mention thatboth qnrB and qnrS genes were detected simultaneously in 5% of ourK. pneumoniaeisolates. That was in accordance with other studies that reported simultaneous detection of both genes in 2.5% and1.8% of K. pneumoniaeisolates.28,37
In this study, the qnrA gene was not detected in any of the studied isolates. This was consistent with two studies in Egypt and other studies from Iran, Korea and France.5,11,13,23,27,43 In contrast other Egyptian studies byTohamy et alfound that of the 70 MDR Gram negative bacilli isolates 2 (2/70, 2.8%) isolates carried qnrAgene26 and Esmat and Mohamed 44 who detected qnrAin 11 % K. pneumoniae isolates. In Kuwait Vali et al45 retorted that only one isolate out of 173 K. pneumonia isolates had qnrAand another Japanese study reported that the prevalence of the qnrA gene was 0.8% in E. coli[36]. This worldwide relatively low prevalence may explain its absence among our isolates.
Qnr genes were detected mainly in blood culture samples (47.2%). Whereas, Peymaniet al[5]reported that qnr-positive isolates were mostly recovered from urine 42.9% followed by trachea secretion 36.7%, wound swabs 12.2%, and were least in blood culture specimens (2%)This may be explained by the difference in the number of each type of specimens in the two studies; as blood was the majorspecimen in the current study whereas urine was the predominant one in the later. Again this figure confirms serious consequences of these resistant strains.
In the present study, all the tested isolates (100%) were resistant to at least one quinolone, while 96.7 % were resistant to all of them. All of the 40 (100%) isolates of K. pneumonia, the most frequently isolated species, were resistant to nalidixic acid, ciprofloxacin and levofloxacin. These results are in accordance with the findings of study in Portugal that reported 100% resistance of K. pneumoniae isolates to nalidixic acid and ciprofloxacin.21
Regarding quinolones susceptibility within qnrS positive isolates; all isolates showed resistance to all quinolones except one E. cloacae isolate that was sensitive only to levofloxacin. Similar findings by Jlili et al[28]who reported that of the 23 qnr-positive K. pneumoniae isolates, 95.6%, 100%, and 69.5% were non-susceptible to nalidixic acid, ciprofloxacin, levofloxacin, respectively. These minor fluctuations in sensitivity patterns to various quinolones could be explained by Jacoby,46 who reported that quinolone susceptibility is decreased by PMQR however, this resistance is not detected in the clinical level, but it enhances other resistance mechanisms and aid further mutations to higher levels of resistance. Also, other mechanisms of resistance rather than PMQR may be involved. In our study; 40% (24/60) of our isolates were negative to qnr genes. These isolates probably had other mechanisms of resistance that were not investigated in this study. Qnr positive isolates infections may consequently augment the selection of these resistant mutants and increase thespread of this type of resistance.
The alliance of quinolone resistance with resistance to other antibiotics especially aminoglycosides was analyzed in this study.Among qnr positive isolates, 97% were resistant to amikacin. The qnr gens presence were significantly correlated with amikacin (p<0.001).That was similar to a Korean study found that allqnrpositive clinical isolates were resistant to amikacin.38 Another Iranian study byMajlesi et al.11 stated that the prevalence of plasmid-mediated quinolone resistance due to the qnr and aac(6′)-Ib-cr genes was high among quinolone-resistant clinical isolates of Enterobacteriaceae where only 41% of quinolone-resistant clinical isolates were positive for PMQR genes.This could be attributed to collocate of PMQR genes with other resistance determinants within transposons and/or integrons in multidrug resistance plasmids. Quinolones and aminoglycosides can act as potential coselectors for antimicrobial resistance. Also the presence of widely prevalent aac (6’ )-Ib-cr47 which confers resistance to kanamycin, tobramycin, netilmicin, amikacin, and ciprofloxacin, although it was not attempted in the current study, may explain the associated high aminoglycosides resistance.48
Conclusion
The prevalence of quinolone resistant Gram negative bacilli clinical isolates in the current study was high. The mostly detected PMQR gens were qnrS followed by qnrB. Gram-negative bacteria expressed high antibiotic resistance common antibiotics lead to loss of convenience for treatment of many infections. More knowledge about trends of antibiotic resistance and mechanisms of resistance is required to reduce the risk of antibiotic treatment failure. The overuse of quinolone has led to increased resistance, making them less effective.
Conflict of interest
There is no conflict of interest.
Acknowledgments
The author(s) received no specific funding for this work.
References
- Cerceo E., Deitelzweig S. B., Sherman B. M., Amin A. N. Multidrug-resistant gram-negative bacterial infections in the hospital setting: Overview, implications for clinical practice and emerging treatment options. Microb Drug Resist. 2016;22(5):412-431.
CrossRef - Agyepong N., Govinden U., Essack S. Y., Owusu-Ofori A. Multi drug-resistant gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrob. Resist. Infect. Control. 2018;7(1).
CrossRef - Nithya N., Remitha R., Faisal M., Kumar P. R. M., Jayasree P. R. Analysis of beta-lactamases, bla NDM-1 phylogeny & plasmid replicons in multidrug-resistant Klebsiella from a tertiary care centre in south India. Indian J Med Res. 2017;146:38-45.
CrossRef - Köck R., Siemer P., Esser J., Kampmeier S., Berends M. S., Glasner C., Arends J. P., Becker K., Friedrich A. W. Defining Multidrug Resistance of Gram-Negative Bacteria in the Dutch-German Border Region-Impact of National Guidelines. Microorganisms. 2018;6(1).
- Peymani A., NaserpourFarivar T., Nikooei L., Najafipour R., Javadi A., Pahlevan A. A. Emergence of plasmid-mediated quinolone-resistant determinants in Klebsiellapneumoniae isolates from Tehran and Qazvin provinces. Iran. J Prev Med Hyg. 2015;56(2):61-65.
- Shahcheraghi F., Nobari S., Ghezelgeh F. R., Nasiri S., Owlia P., Nikbin V. S.,Fooladi A. A. I. First report of New Delhi metallo-beta-lactamase-1-producing Klebsiellapneumoniae in Iran. Microb Drug Resist. 2013;19(1):30-36.
CrossRef - Briales A., Rodríguez-Martínez J. M., Velasco C., de Alba P. D., Rodriguez-Bano J., Martinez-Martinez L., Pascual A. Prevalence of plasmid-mediated quinolone resistance determinants qnr and aac (6′)-Ib-cr in Escherichia coli and Klebsiellapneumoniae producing extended-spectrum β-lactamases in Spain. Int J Antimicrob Agents. 2012;39(5):431-434.
CrossRef - Alikhani M. Y., Hashemi S. H., Aslani M. M., Farajnia S. Prevalence and antibiotic resistance patterns of diarrheagenicEscherichia coli isolated from adolescents and adults in Hamedan, Western Iran. Iran J Microbiol. 2013;5(1):42-47.
- Amin M., Dibachi S., Shahin M. Prevalence of class 1 integrons and plasmid-mediated qnr-genes among Enterobacter isolates obtained from hospitalized patients in Ahvaz, Iran. Infez Med. 2017; 25(4):351-357.
- Ranjbar R., Farahani O. The prevalence of plasmid-mediated quinolone resistance genes in Escherichia coli isolated from hospital wastewater sources in Tehran, Iran. Iran J Public Health. 2017;46(9):1285-1291.
- Majlesi A., Kakhki R. K., MozaffariNejad A. S., Mashouf R. Y., Roointan A., Abazari M., Alikhani M. Y. Detection of plasmid-mediated quinolone resistance in clinical isolates of Enter obacteriaceae strains in Hamadan, West of Iran. Saudi J Biol. Sci. 2018;25(3):426-430.
CrossRef - Amin A. K., Wareham D. W. Plasmid-mediated quinolone resistance genes in Enter obacteriaceae isolates associated with community and nosocomial urinary tract infection in East London, UK. Int J Antimicrob Agents. 2009;34(5):490-491.
CrossRef - El-Badawy M. F., Tawakol W. M., El-Far S. W., Maghrabi I. A., Al-Ghamdi S. A., Mansy M. S., Ashour M. S., Shohayeb M. M. Molecular identification of aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance genes among Klebsiellapneumoniae clinical isolates recovered from Egyptian patients. Int J Microbiol. 2017;1-12.
CrossRef - Strahilevitz J., Jacoby G. A., Hooper D. C., Robicsek A. Plasmid-Mediated Quinolone Resistance: a Multifaceted Threat. Clin. Microbiol Rev. 2009;22(4):664-689.
CrossRef - Padmini N., Ajilda A. A. K., Sivakumar N., Selvakumar G. Extended spectrum b-lactamase producing Escherichia coli and Klebsiellapneumoniae: critical tools for antibiotic resistance pattern. J Basic Microbiol. 2017;57(6):460-470.
CrossRef - Green M. R., Sambrook J. Molecular cloning a laboratory manual. Cold Spring Harbor, N.Y. Cold Spring Harbor Laboratory Press. 2012.
- M100-S28 performance standards for antimicrobial susceptibility testing; Twenty-eight informational supplement. 2018.
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST a new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402.
CrossRef - Mammeri H., De Loo M. V., Poirel L., Martinez-Martinez L., Nordmann P. Emergence of Plasmid-Mediated Quinolone Resistance in Escherichia coli in Europe. Antimicrob Agents Chemother. 2005;49(1):71-76.
CrossRef - Cattoir V., Poirel L., Rotimi V., Soussy C. J., Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enter obacterial isolates. J Antimicrob. Chemother. 2007;60(2):394-397.
CrossRef - Ferreira S., Toleman M., Ramalheira E., Da Silva G. J., Walsh T., Mendo S. First description of Klebsiellapneumoniae clinical isolates carrying both qnrA and qnrB genes in Portugal. Int J Antimicrob Agents. 2010;35(6):584-586.
CrossRef - Herrera-Leâon S., Gonzâalez-Sanz R., Herrera-Leâon L., Echeita M. A. Characterization of multidrug-resistant Enterobacteriaceae carrying plasmid-mediated quinolone resistance mechanisms in Spain. J Antimicrob. Chemother. 2011;66(2):287-290.
CrossRef - Yang H., Duan G., Zhu J., Zhang W., Xi Y., Fan Q. Prevalence and characterisation of plasmid-mediated quinolone resistance and mutations in the gyrase and to poisomerase IV genes among Shigella isolates from Henan, China, between 2001 and 2008. Int J Antimicrob Agents. 2013; 42(2):173-177.
CrossRef - Jiang X., Li J., Zhang Y., Yan H., Wang Y., Shi L., Zhou L . Detection of plasmid-mediated quinolone resistance determinants and qnrS expression in Enterobacteriaceae clinical isolates. J Infect Dev. Ctries. 2014;8(12):1625-1629.
CrossRef - Gamal D., Fernández-Martínez M., Salem D., El-Defrawy I., Montes L. Á., Ocampo-Sosa A. A., Martínez-Martínez L. Carbapenem-resistant Klebsiellapneumoniae isolates from Egypt containing blaNDM-1 on Inc R plasmids and its association with rmt F. Int J Infect Dis. 2016;43:17-20.
CrossRef - Tohamy S. T., Aboshanab K. M., El-Mahallawy H. A., El-Ansary M. R., Afifi S. S. Prevalence of multidrug-resistant Gram-negative pathogens isolated from febrile neutropenic cancer patients with bloodstream infections in Egypt and new synergistic antibiotic combinations. Infect Drug Resist. 2018;11:791-803.
CrossRef - Helmy O. M., Kashef M. T. Different phenotypic and molecular mechanisms associated with multidrug resistance in Gram-negative clinical isolates from Egypt. Infect Drug Resist. 2017;10: 479-498.
CrossRef - Jlili N. E. H., Réjiba S., Smaoui H., Guillard T., Chau F., Kechrid A., Cambau E. Trend of plasmid-mediated quinolone resistance genes at the Children’s Hospital in Tunisia. J Med Microbiol. 2014; 63(2):195-202.
CrossRef - Hamed S. M., Aboshanab K. M., El-Mahallawy H. A., Helmy M. M., Ashour M. S., Elkhatib W. F. Plasmid-mediated quinolone resistance in Gram-negative pathogens isolated from cancer patients in Egypt. Microb Drug Resist. 2018. https://doi.org/10.1089/mdr. 2017.0354.
- Orsini J., Mainardi C., Muzylo E., Karki N., Cohen N., Sakoulas G. Microbiological profile of organisms causing bloodstream infection in critically ill patients. J Clin Med Res. 2012;4(6):371-377.
CrossRef - Marando R., Seni J., Mirambo M. M., Falgenhauer L., Moremi N., Mushi M. F.,Kayange N., Manyama F., Imirzalioglu C., Chakraborty T., Mshana S. E. Predictors of the extended-spectrum-beta lactamases producing Enterobacteriaceae neonatal sepsis at a tertiary hospital Tanzania. Int J Med Microbiol. 2018;6:12. DOI: 10.1016/j.ijmm.
- Charfi K., Grami R., Jeddou A. B., Messaoudi A., Mani Y., Bouallegue O., Boujaafar N., Aouni M., Mammeri H., Mansour W. Extended-spectrum β-lactamases and plasmid-mediated quinolone resistance in enterobacterial clinical isolates from neonates in Tunisia. Microb. Pathog. 2017;110: 184-188.
CrossRef - Bouchakour M., Claude J. D. P. G., Timinouni M., Zerouali K., Mdaghri N. E., Amarouch H., Courvalin P. Plasmid-mediated quinolone resistance in expanded spectrum beta lactamase producing enterobacteriaceae in Morocco. J Infect Dev. Ctries. 2010;4(12):799-803.
- Poulou A., Grivakou E., Vrioni G., Koumaki V., Pittaras T., Pournaras S., Tsakris A. Modified CLSI extended-spectrum b-lactamase (ESBL) confirmatory test for phenotypic detection of ESBLs among Enterobacteriaceae producing various b-lactamases. J Clin. Microbiol. 2014;52(5):1483-1489.
CrossRef - Richter S. N., Frasson I., Bergo C., Manganelli R., Cavallaro A., Palù G .Characterisation of qnr plasmid-mediated quinolone resistance in Enterobacteriaceae from Italy: association of the qnrB19 allele with the integron element ISCR1 in Escherichia coli. Int J Antimicrob Agents. 2010;35(6):578-583.
CrossRef - Okade H., Nakagawa S., Sakagami T., Hisada H., Nomura N., Mitsuyama J., Yamagishi Y., Mikamo H .Characterization of plasmid-mediated quinolone resistance determinants in Klebsiella pneumoniae and Escherichia coli from Tokai, Japan. J Infect Chemother. 2014;20(12):778-783.
CrossRef - Yang H., Chen H., Yang Q., Chen M., Wang H. High prevalence of plasmid-mediated quinolone resistance genes qnr and aac (6′)-Ib-cr in clinical isolates of Enterobacteriaceae from nine teaching hospitals in China. Antimicrob Agents Chemother. 2008;52(12):4268-4273.
CrossRef - Tamang M. D., Seol S. Y., Oh J. Y., Kang H. Y., Lee J. C., Lee Y. C. Plasmid-Mediated Quinolone Resistance Determinants qnrA, qnrB, and qnrS among Clinical Isolates of Enterobacteriaceae in a Korean Hospital. Antimicrob Agents Chemother. 2008;52(11):4159-4162.
CrossRef - Karah N., Poirel L., Bengtsson S., Sundqvist M., Kahlmeter G., Nordmann P., Sundsfjord A., Samuelsen Ø. Norwegian Study Group on PMQR. Plasmid-mediated quinolone resistance determinants qnr and aac (6′)-Ib-cr in Escherichia coli and Klebsiella from Norway and Sweden. Diagn. Microbiol Infect Dis. 2010;66(4):425-431.
CrossRef - Poirel L., Leviandier C., Nordmann P. Prevalence and Genetic Analysis of Plasmid-Mediated Quinolone Resistance Determinants QnrA and QnrS in Enterobacteriaceae Isolates from a French University Hospital. Antimicrob Agents Chemother. 2006;50(12):3992-3997.
CrossRef - Hassan W. M., Hashim A., Domany R. A. A. Plasmid mediated quinolone resistance determinants qnr, aac (6′)-Ib-cr, and qep in ESBL-producing Escherichia coli clinical isolates from Egypt. Indian J Med Microbiol. 2012;30(4):442-447.
CrossRef - Jeong H. S., Bae I. K., Shin J. H., Jung H. J., Kim S. H., Lee J. Y., Oh S. H., Kim H. R., Chang C. L., Kho W. G., Lee J. N . Prevalence of plasmid-mediated quinolone resistance and its association with extended-spectrum beta-lactamase and AmpC beta-lactamase in Enterobacteriaceae. Korean J Lab Med. 2011;31(4):257-264.
CrossRef - Crémet L., Caroff N., Dauvergne S., Reynaud A., Lepelletier D., Corvec S. Prevalence of plasmid-mediated quinolone resistance determinants in ESBL Enterobacteriaceae clinical isolates over a 1-year period in a French hospital. Patho. lBiol. 2011;59(3):151-156.
CrossRef - Esmat M. M., Mohamed H. H. Quinolone resistance among extended-spectrum β-lactamases Producing Klebsiellapneumoniae in Sohag university hospital, Upper Egypt. EJMM. 2016;25(1): 69-76.
CrossRef - Vali L., Dashti A. A., Jadaon M. M., El-Shazly S. The emergence of plasmid mediated quinolone resistance qnrA 2 in extended spectrum β-lactamase producing Klebsiellapneumoniae in the Middle East. J Pharm Sci. 2015;23(1):34.
- Jacoby G. A. Study of Plasmid-Mediated Quinolone Resistance in Bacteria. In DNA Topoisomerases. Humana Press, New York, NY. 2018;317-325.
CrossRef - Fatima E. L., Meriem E. L., Said B. Plasmid mediated quinolones resistance ESBL-Enterobactériaceae in Moroccan. Pharmaceut Anal Acta. 2012;15:1-4.
- Wen Y., Pu X., Zheng W., Hu G. High Prevalence of Plasmid-Mediated Quinolone Resistance and IncQ Plasmids Carrying qnrS2 Gene in Bacteria from Rivers near Hospitals and Aquaculture in China. PloS one. 2016;11(7).
CrossRef







