Sonali Nandish Manoli1, Anand Raj Ulle1 , N. M. Nandini2 and T. S. Rekha2
, N. M. Nandini2 and T. S. Rekha2
1Department of Information Science and Engineering, Sri Jayachamarajendra College of Engineering, Mysuru.
2Department of Pathology, JSS Medical College, Affiliated to JSS University Mysuru.
Corresponding Author E-mail: sonalinandish@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1544
Abstract
In this paper, we propose a novel method to classify Breast Lesions based on minute changes in the cell and nuclear features of the cell. It is important to note these changes as they play a significant role in diagnosis and the line of treatment by an oncologist. To overcome the problem of inter-observer variability the method of scoring is used to grade the lesions considered for the study. We have used the Modified Masood Score and designed an algorithm which classifies a given breast lesion into 6 classes namely Benign, Intermediate class-1,Intermediate class-2, Malignant class-1,Malignant class-2 and Malignant class-3. We have developed a sensitive model using the feed-forward neural network and Pattern Network to achieve the above objective. The Rank of the features is observed using ReliefF Algorithm.
Keywords
Computer Aided Diagnosis; Feed-Forward Neural Network; Machine Learning, Modified Masood Score; Neural Networks; Pattern Network; Performance Function and Training Function
Download this article as:| Copy the following to cite this article: Manoli S. N, Ulle A. R, Nandini N. M, Rekha T. S. Classification of Breast Lesions using Modified Masood Score and Neural Network. Biomed Pharmacol J 2018;11(3). |
| Copy the following to cite this URL: Manoli S. N, Ulle A. R, Nandini N. M, Rekha T. S. Classification of Breast Lesions using Modified Masood Score and Neural Network. Biomed Pharmacol J 2018;11(3). Available from: http://biomedpharmajournal.org/?p=21872 |
Introduction
Breast lesion is an extra growth or lump in the tissues of the Breast.1 There is a necessity to diagnose the condition of the breast lesion because it is estimated that 13.4 % of the women born today will be diagnosed with cancer at some stage in their life.2 For better diagnosis the intervention of machines is necessary as they help in removing human errors caused by fatigue, oversight and inter-observer variability. To develop such a system computer-aided diagnosis plays a major role. For breast cancer detection the earliest systems where developed using supervised machine learning approaches by classifying the lesion into Benign(Non-Cancerous) and Malignant (cancerous) condition.3-4 To improve the detection of breast lesion it is also important to look into samples which progress from the benign lesion towards malignancy. It gives rise to intermediate condition which is neither Benign nor Malignant.
The detection of such lesions can be achieved under cytology by a cost effective method called as the Fine Needle Aspiration Cytology.5 Under Medical domain, grading system has been introduced to give a range for the observation of the characteristic features and if a given sample falls under this range it is classified into one condition of the breast lesion.
Shahla Masood in5 has proposed multi-class classification proposing the Masood score. The score has been modified and validated to improve the grading system for more accurate diagnosis.6-8 Among the two scoring systems namely Masood Score and the Modified Masood Score it is proved that Modified Masood Score is better for more accurate classification of breast lesions.9 Hence Modified Masood Score has been considered in the study.
From10 we can observe that the Malignant condition can be classified further into grade -I , grade-II, grade-III. This change under malignant condition occurs to due to minor changes in the characteristic features. Each condition has a different line of treatment but if untreated it may cause progress of the disease, reoccurrence or even death.The machine which is sensitive cannot be effectively obtained using conventional classifiers when the data is divided into train and test data set.
To handle this problem, sensitive machine is required for classification. Hence we have considered a Feed Forward Neural Network and Pattern Network where both have one hidden layer in our study. The network can be trained to classify the input features to a particular class by setting the targets as outputs.11-12 In our system we have used linear regression to perform the classification of the breast lesion samples. To observe the classification accuracy various algorithms to train the neural network are used namely: Levenberg-Marquardt, Bayesian Regularisation, BFGS Quasi newton, Reselient Back-Propagation, Scaled-conjugate Gradient, Conjugate Gradient with powell restart, Conjugate Gradient with bealle restart, Fletcher-Powell Conjugate Gradient, Polak-Ribiére Conjugate Gradient, One Step Secant, Variable Learning Rate Gradient Descent, Gradient Descent with Momentum, Gradient Descent.13-15 Each of the above methods are further trained using cross entropy , sum of absolute error, Mean of absolute error, sum of squared error, mean of squared error.16 The Rank of the features is obtained using ReliefF Agorithm.17
Implementation Methodlogy
Dataset
For doing the data analysis using the Modified Masood Score, the samples were obtained from the Department of Pathology at JSS Hospital, JSS Medical College, Mysore. The study uses 321 such samples in which 122 samples belong to Benign or Non-Proliferative Breast Disease class, 64 samples belong to Intermediate class-1 or Proliferative Breast Disease without atypia class, 25 samples belong to Intermediate class -2 or Proliferative Breast Disease with Atypia class, 55 samples belong to Carcinoma class-1, 40 samples belong to Carcinoma class-2, 15 samples belong to Carcinoma class-3. Here both the intermediate classes indicate a condition where the breast lesion is moving towards cancerous condition and all the carcinoma classes indicate malignant condition or cancer which requires accurate treatment based on the severity of the disease indicated by class.
Experimentation and Results
The classification of the samples has been performed by using the feed-forward neural network (FNN). To test the accuracy of classification the samples have been considered in the ratio 70:30 as train : test. The feed-forward neural network is built using the input layer , the hidden layer and the output layer. In the input layer the six characteristic features with the scores are given as input. The weight and bias perform the activation function based on the sigmoid operation using neurons in the hidden layer. During experimentation it is observed that the neural network performed the best fit using 30 neurons for training in the hidden layer. The system performed under-fitting when the neurons used for training were lesser than 30 and it performed over-fitting when the neurons used for training were above 30. Hence the overall accuracy would decrease in both under- fitting and over-fitting. So 30 neurons were fixed to train the network using the feed-forward neural network. To enhance the accuracy of performance of the network, twelve different training algorithms were used as training functions. The best accuracy was given by the Bayesian Regularisation function with the accuracy of 87.53 %.
To further enhance the network performance, Pattern- Neural Network with the same features, train: test ratio and neurons was used to perform classification. Under this network the performance of the network was measured by considering five performance measures.
The results obtained when these various training algorithms and performance measures were used is as shown in Figure 1.
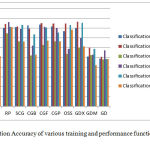 |
Figure 1: Classification Accuracy of various training and performance functions in FNN and PNN
|
In the above Figure, Y Axis indicates the Classification Accuracy and the X-Axis indicates the training functions denoted as LM ,BR, BFG, RP, SCG, CGB, CGF, CGP, OSS, GDX, GDM, GD to indicate twelve training functions Levenberg-Marquardt, Bayesian Regularisation, BFGS Quasi newton, Reselient Back-Propagation,Scaled-conjugate Gradient , Conjugate Gradient with powell restart, Conjugate Gradient with bealle restart, Fletcher-Powell Conjugate Gradient, Polak-Ribiére Conjugate Gradient, One Step Secant, Variable Learning Rate Gradient Descent, Gradient Descent with Momentum and Gradient Descent respectively.
The five performance measures used to observe the performance of each training function are Cross-Entropy(CE), Sum of Absolute Error(SAE) ,Sum of Squared Error(SSE), Mean Of Absolute Error(MAE), Mean of Squared Error(MSE).
Here FNN Indicates Feed-Forward Neural Network and PNN Indicates Pattern Network.
Finally based on the above target functions and performance parameters used for activation, the neuron is fired to the target in the output layer based on which is the most probable class, the sample belongs to. The difference between the expected output target and the observed output target gives the error rate in performance of the network or in otherwords the accuracy of the system. The highest accuracy obtained is using Bayesian regularisation with accuracy of 87.53 % in the feed forward neural network. An accuracy of 88.44% is obtained using pattern network with Bayesian Regularisation as the training function and with cross-entropy as the performance parameter.
The rank importance of each feature is obtained using ReliefF Algorithm. It is as shown in the table below :
Table 1: Six features used in the study and rank obtained to show the most and least significant feature.
| Features 1 to 6 initially | Cellular Arrangement | Cellular Pleomorphism | Myoepithelial cells | Anisonucleosis | Nucleoli | Chromatin Clumping |
| Rank | 6 | 2 | 4 | 1 | 5 | 3 |
| Features 1 to 6 according to Rank | Chromatin Clumping | Cellular pleomorphism | Anisonucleosis | Cellular Arrangement | Nucleoli | Myoepithelial cells |
So from the above table we can understand that chromatin clumping is the most significant feature and Myoepitheial cells are the least significant feature to classify breast lesions.
Conclusion
In this paper , an automated method of classifying breast lesions into six classes based on Modified Masood Score is presented. This system overcomes the problem of classification in conventional classifiers when samples in a particular class are very less, It is efficient to classify samples even though the number of samples present in each class varies greatly with respect to another class. The system is chosen because it is simple and cost-effective to categorize the breast lesions. The system is also sensitive to minor changes in the scores of characteristic features which help an oncologist to give the most effective dosage of treatment for early recovery. The rank of features has been approved by the pathologists involved in the study. It is an efficient step towards precise treatment for the cancer patient. A system which classifies the image of a breast lesions with better accuracy and sensitivity is being developed.
References
- NCI Homepage, http://www.cancer.gov/types/breast/patient/breast-treatment-pdq#section all, last accessed .2017/9/14.
- Vig L. Comparative Analysis of Different Classifiers for the Wisconsin Breast Cancer Dataset. Open Access Library Journal. 2014;1(660):1-7.
CrossRef - Wolberg W.H, Mangasrin O.L. Multisurface Method of pattern separation for medical diagnosis applied to breast cytology. Proc.Natl.Acad.Sci.USA. 1990;87(23):9193-9196.
CrossRef - Yueqiao Z, Hong R. Meta-analysis of diagnostic accuracy of magnetic resonance imaging and mammography for breast cancer. Journal of cancer research and therapuatics. 2017;13(5):862-868.
CrossRef - Masood S. Cytomorphology of Fibrocystic Change, High-Risk Proliferative Breast Disease and Premalignant Breast Lesions.Clin.Lab.Med. 2005;25(4):713-731.
CrossRef - Ranjan A.M, Iyer V.K, Kapila K, Verma K. Value of scoring system in classification of Proliferative Breast Disease on Fine Needle Aspiration Cytology.Indian.J.Pathol.Microbiol. 2006;49(3):334-340.
- Nandini N.M,Rekha T.S,Manjunath G.V. Evaluation of Scoring System in Cytological diagnosis and management of breast lesion with review of literature.Indian.J.Cancer. 2011;48(2):240-245.
CrossRef - Sheeba D, Sugumar C. Palpable Breast Lesons-Cytomorphological Analysis and Scoring System with Histopathological Correlation. IOSR-JDMS. 2016;15(10):25-29.
CrossRef - Krishna S.C, Moothiringode S.C. Evaluation of Masood’s and Modified Masood’s Scoring System in the Cytological Diagnosis of Palpable Breast Lump Aspirates .J.Clin.Diagn.Res. 2017;11(4):06-10.
- Rekha T.S , Nandini N.M, Dhar M. Expansion of Masood’s cytologic Index for Breast Carcinoma and its validity.Journal of Cytology. 2013;30(4):233-236.
CrossRef - Sharma B , Venugopalan K. Hematoma Classification in Brain CT Images. IOSR Journal of Computer Engineering (IOSR-JCE). 2014;16(1):31-35.
CrossRef - Abdolmaleki P, Buadu L.D, Murayama S, Murakami J, Hashiguchi N, Yabuuchi H,Masuda K. Neural network analysis of breast cancer from MRI findings. Radiat.Med. 1997;15(5):283-293.
- Mishra S,Prusty R, Kumar P.H. Analysis of Levenberg-Marquardt and Scaled Conjugate gradient training algorithms for artificial neural network based LS and MMSE estimated channel equalizers.International Conference Man and Machine Interfacing (MAMI). 2015.
CrossRef - Krzysztof K.C. Convergence and efficiency of subgradient methods for quasiconvex minimization. Mathematical Programming, Springer. 2001;90(1):1–25.
CrossRef - Ya-xiang Y. Step-sizes for the gradient method, AMS/IP Studies in Advanced Mathematics. Providence, RI. American Mathematical Society. 1999;42(2):785.
- Willmott C.J, Matsuura K. On the use of dimentioned measures of error to evaluate the performance of spatial interpolators. International Journal of Geographical Information Science. 2006;20:89–102.
CrossRef - Durgabai R.P.L. Feature Selection using ReliefF Algorithm. International Journal of Advanced Research in Computer and Communication Engineering. 2014;3(10):8215-8216.
CrossRef








