Hussein O. M. Al-Dahmoshi1 , Noor S. Al-Khafaji2, Ahmed Abdulzahra Jeyad3, Hasanain Khaleel Shareef4 and Rafah F. Al-Jebori5
, Noor S. Al-Khafaji2, Ahmed Abdulzahra Jeyad3, Hasanain Khaleel Shareef4 and Rafah F. Al-Jebori5
1,2Biology Dept., College of Science-University of Babylon, Hilla-Iraq.
3Al-Sadiq Teaching Hospital, Babylon Health Directorate-Ministry of Health-Iraq.
4Biology Dept., College of Science for Women -University of Babylon, Hilla-Iraq.
5Laboratory Devision, Babylon Health Directorate-Ministry of Health-Iraq.
Corresponding Author E-mail: dr.dahmoshi83@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1439
Abstract
Wound infections regards one of the most common infections encountered in hospital records. Pseudomonas aeruginosa regard the 3rd common pathogen among healthcare-related infections. Their ability to adapt to different conditions and presence of pool of virulence factors may render their infections delay in healing. During a period of six months 114 wound swabs were collected and inoculated on Pseudomonas chromogenic agar and then Pseudomonas aeruginosa isolated confirmed by PCR using specific primer for 16S rDNA gene of Pseudomonas aeruginosa. Molecular investigation of some virulence factor like ExoA, OprL, OprI, LasI and LasB were performed using a sets of specific primer pairs. The results revealed that only 26 (22.8%) isolates were Pseudomonas aeruginosa and the coexistence of more than one virulence factors within the same isolates was also recorder. OprI and LasB were most common followed by LasI, ExoA and OprL. Occurrence of virulence factor genes were 12(46.15%) for exoA, oprL was 11(42.3%), oprI was 22(84.61%), lasI was 14(53.84%) and lasB was 18(69.23%). Results of this study can lead us to conclude that P. aeruginosa have an arrays of virulence traits via which can adapt to different conditions and so cause a wide-ranging of hard to cured infections and the delay in healing and worseness degree may be attributed to owning multivirulence factors.
Keywords
Aeruginosa; ExoA; LasI; LasB P. Virulence;
Download this article as:| Copy the following to cite this article: Al-Dahmoshi H. O. M, Al-Khafaji N. S, Jeyad A. A, Shareef H. K, Al-Jebori R. F. Molecular Detection of Some Virulence Traits Among Pseudomonas aeruginosa Isolates, Hilla-Iraq. Biomed Pharmacol J 2018;11(2). |
| Copy the following to cite this URL: Al-Dahmoshi H. O. M, Al-Khafaji N. S, Jeyad A. A, Shareef H. K, Al-Jebori R. F. Molecular Detection of Some Virulence Traits Among Pseudomonas aeruginosa Isolates, Hilla-Iraq. Biomed Pharmacol J 2018;11(2). Available from: http://biomedpharmajournal.org/?p=20508 |
Intriduction
Pseudomonas aeruginosa is a momentous life-intimidating, hospital pathogen that play a noticeable role in wound infections of burned patients.1,2 It is success can be attributed to wide arrays of virulence factors which leads to adaptation and withstand for different inconvenient niches. Burn injuries stays one of the utmost public forms of trauma that push a major community health problem internationally. Skin compile a physical barricade against invasive microbes and the infections resulted from burn is common when this barrier breached. It is the supreme cause of burn wound infections.3,4,5,6 The notable aptitude of P. aeruginosa to acclimatize and flourish among varied environments due to its extensive inherited adaptability and its intrinsic and acquired resistance to different antibiotics, which contributes meaningfully to its possible pathogenicity[7,8]. Wound-associated infections are frequently hard to treat due to various bacterial pathogens.9,10,11,12,13
P. aeruginosa have an arrays of virulence factors that frustrate host defenses and make direct tissue’s harm or rise bacterium’s affordability. The most important virulence factors of P. aeruginosa includes Exotoxin A (ETA), outer membrane associated protein L and I and quorum-sensing determinant system.14 Exotoxin A represent the main dangerous virulence factor produced by P. aeruginosa.15 OprL and OprI are peptidoglycan-related outer membrane proteins that fetter the outer membrane to the principal peptidoglycan. Lipoprotein of outer membrane lipoprotein (OprL) concerned in purge transporting systems leading to disturbing cell penetrability.16 They are accountable for intrinsic resistance to antiseptics and antibiotics. Quorum-sensing (QS) system permits bacteria to respond to their community mass by harmonized adaptable their gene expression patterns. It is vital for the countenance of a series of virulence traits in addition to biofilm development.17,18,19,20
The current study aimed to investigate the occurrence of exoA, oprL, oprI, lasI and lasB gene among P. aeruginosa isolated from wound infection in Hilla-Iraq.
Materials And Methods
Samples
Wounds of 114 burned patients were swabbed from patients admitted to Al-Hilla teaching hospitals, Hilla-Iraq during a period of 6 months. All swabs were firstly cultivated on Pseudomonas chromogenic agar plate (Condalab/Spain) and then confirmed as Pseudomonas aeruginosa by amplification of 16S rDNA gene using specific primer pairs.
DNA Extraction
Preparation of bacterial samples were performed according to Al-Dahmoshi (2017)[6] and then the steps of protocol of Mini DNA extraction kit (Favorgen/Taiwan) were followed. The extracted DNA checked using gel electrophoresis (0.7%) and then stained with safe DNA stain, RedSafe (IntronBio/Korea) and then visualized using Gel Documentation (Vilber/France).
Preparation of Primer and PCR
A working solution of primer pair were prepared by addition of nuclease free water (New England Biolabs/UK) to the lyophilized primer pairs (Realgene/China) to get 10pmole/μl concentration. A reaction mixture of 25 μl were prepared using OneTaq Quick-Load 2X Master Mix. The primer sequence and PCR conditions were mentioned in the table 1 and 2 respectively.
Table 1: Show primer pairs sequence and amplicon size.
| Primer Name | Sequence (5՛-3՛) | Amplicon (bp) | References |
| PA-SS-F | GGGGGATCTTCGGACCTCA | 956 | [21] |
| PA-SS-R | TCCTTAGAGTGCCCACCCG | ||
| ETA-F | GACAACGCCCTCAGCATCACCA | 397 | [22] |
| ETA-R | CGCTGGCCCATTCGCTCCAGCG | ||
| oprL-F | ATG GAAATGCTGAAATTCGGC | 504 | [23] |
| oprL-R | CTTCTTCAGCTCGACGCGACG | ||
| oprI-F | ATGAACAACGTTCTGAAATTCTCTGCT | 249 | [23] |
| oprI-R | CTTGCGGCTGGCTTTTTCCAG | ||
| LasI-F | CGTGCTCAAGTGTTCAAGG | 295 | [24] |
| LasI-R | TACAGTCGGAAAAGCCCAG | ||
| lasB-F | TTCTACCCGAAGGACTGATAC | 153 | [24] |
| lasB-R | AACACCCATGATCGCAAC |
Table 2: Show the PCR. conditions.
| Gene | Ini. Denat. | Denat. | Anneal. | Exten. | Final. Exten. | Reference | ||
| 16S rDNA | 95°C.
2 min. |
95°C.
30 sec. |
61°C.
30 sec. |
72°C.
100 sec. |
72°C.
5 min. |
Current study | ||
| # of Cycles | 1 | 30 | 1 | |||||
| exoA | 95°C.
2 min. |
95°C.
30 sec. |
64.6°C.
30 sec. |
72°C.
40 sec. |
72°C.
5 min. |
Current study | ||
| # of Cycles | 1 | 30 | 1 | |||||
| oprL | 95°C.
2 min. |
95°C.
30 sec. |
60.7°C.
30 sec. |
72°C.
30 sec. |
72°C.
5 min. |
Current study | ||
| # of Cycles | 1 | 30 | 1 | |||||
| oprI | 95°C.
2min. |
95°C.
30 sec. |
60.7°C.
30 sec. |
72°C.
30 sec. |
72°C.
5 min. |
Current study | ||
| # of Cycles | 1 | 30 | 1 | |||||
| lasI | 95°C.
2 min. |
95°C.
30 sec. |
56.7°C.
30 sec. |
72°C.
30 sec. |
72°C.
5 min. |
Current study | ||
| # of Cycles | 1 | 30 | 1 | |||||
| lasB | 95°C.
2 min. |
95°C.
30 sec. |
54.6°C.
30 sec. |
72°C.
30 sec. |
72°C.
5 min. |
Current study | ||
| # of Cycles | 1 | 30 | 1 | |||||
Results and Discussion
Results of isolation revealed that 39 isolate were Pseudomonas spp. while amplification of 16S rDNA gene showed that only 26 (22.8%) isolates were Pseudomonas aeruginosa (figure 1). The PCR product was 956 bp (figure 2) and this results was in accordance with many local and international studies who found that the isolation percentage of P. aeruginosa were 22-32%.1,8,12,25,26 The highest isolation percentage may be due to the fact that it is the 3rd popular pathogen related with hospital-acquired. infections.25,26
The PCR results of virulence factor occurrence showed that exoA was present among 12(46.15%), oprL was 11(42.3%), oprI was 22(84.61%), lasI was 14(53.84%) and lasB was 18(69.23%) (figure 3). Amplicon of exoA was 397 bp, 504 bp for oprL, 249 bp for oprI, 295 bp for lasI and 153 bp for lasB (figure 4,5,6,7,8)
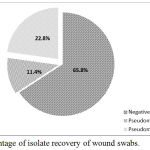 |
Figure 1: Percentage of isolate recovery of wound swabs.
|
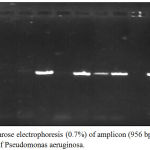 |
Figure 2: Agarose electrophoresis (0.7%) of amplicon (956 bp) of 16S rDNA gene of Pseudomonas aeruginosa.
|
M lane represent 100 bp DNA Marker while the rest lanes represents samples.
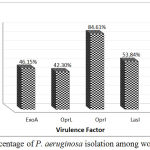 |
Figure 3: Percentage of P. aeruginosa isolation among wound swabs.
|
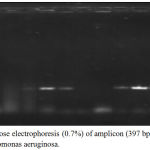 |
Figure 4: Agarose electrophoresis (0.7%) of amplicon (397 bp) of exoA gene of Pseudomonas aeruginosa.
|
M lane represent 100 bp DNA Marker while the rest lanes represents samples.
 |
Figure 5: Agarose electrophoresis (0.7%) of amplicon (504 bp) of oprL gene of P. aeruginosa.
|
M lane represent 100 bp DNA Marker while the rest lanes represents samples.
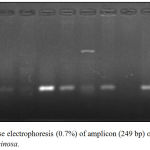 |
Figure 6: Agarose electrophoresis (0.7%) of amplicon (249 bp) of oprI gene of P. aeruginosa.
|
M lane represent 100 bp DNA Marker while the rest lanes represents samples.
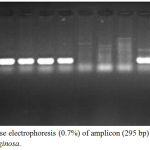 |
Figure 7: Agarose electrophoresis (0.7%) of amplicon (295 bp) of lasI gene of P. aeruginosa.
|
M lane represent 100 bp DNA Marker while the rest lanes represents samples.
 |
Figure 8: Agarose electrophoresis (0.7%) of amplicon (153 bp) of lasB gene of P. aeruginosa.
|
M lane represent 100 bp DNA Marker while the rest lanes represents samples.
Coexistence of more than one virulence factor within the same isolate were recorder and the results displayed that 8/26 have all five virulence factors, 4/26 have four virulence factors, 1/26 have three virulence factors and 5/26 have only two virulence factors (table 3).
Table 3: Show the coexisted virulence factors among isolates.
| Virulence Factor | No. of Isolates |
| ExoA, OprL, OprI, lasI, LasB | 8 |
| ExoA, OprI, lasI, LasB | 2 |
| OprL, OprI, lasI, LasB | 2 |
| OprI, LasI, LasB | 1 |
| OprI, LasB | 2 |
| OprI, LasI | 2 |
| OprL, OprI | 1 |
| Total | 18 |
ExoA responsible for toxigenesis trait of P. aeruginosa while invasiveness achieved by LasB and so the coexistence of ExoA, LasI and LasB let both of mechanism of infection available and increase the degree of wound worseness.27,28,29,30 ExoA had distinct role in hindrance of wound contraction and remedial.15 Both of OprL and OprI have a role in antibiotic resistance via efflux mechanism and alterations of membrane permeability. OprL, OprI and LasI were engaged in antibiotic resistance and biofilm creation leads to many problems concerning treatment of P. aeruginosa infections and make the infection hard to cured.31,32 Bacteria attachment and immune system disruption can be facilitated by LasB. It is a protease (Elastase) that splits collagen, immunoglobulin G and A and complement moreover to destruction of fibronectin to uncover ligands for bacterial adhesion[27]. Nikbin et al., (2012) found that all isolates carried oprI, oprL and lasB genes.16 The presence of ExoA, OprL, OprI, LasI and LasB among P. aeruginosa isolates suppose their linking with different levels of intrinsic virulence and pathogenicity.33,34 The presence of toxins and enzymes genes make them important clinically and environmtally.35
Results of this study can lead us to conclude that P. aeruginosa have an arrays of virulence traits via which can adapt to different conditions and so cause a wide range of hard to cured infections and the delay in healing and worseness degree may be attributed to owning multivirulence factors.
Acknowledgment
I would like to show our gratitude to the staff of advanced microbiology laboratory-College of science, University of Babylon for their assistance.
Conflict of Interest
There is no conflict of interest
Funding Source
The financial support is fully personal.
References
- Al-Ahmadi G.J and Roodsari R.Z. Fast and specific detection of Pseudomonas Aeruginosa from other pseudomonas species by PCR. Annals of burns and fire disasters. 2016;29(4):264.
- Naher H.S, Al-Dahmoshi H.O, Al-Khafaji N.S, AL-Saffar A.K and AL-Saffar H.K. Anti-Pseudomonal Effect of Argan Oil on Pseudomonas aeruginosa Recovered from Burn Patients in Hilla City, Iraq. The Journal of Infectious Diseases. Photon. 2014;113:270-276.
- Church D, Elsayed S, Reid O, Winston B. and Lindsay R. Burn wound infections. Clinical microbiology reviews. 2006;19(2):403-434.
CrossRef - Abdulazeem L, Al-Dahmoshi H.O and Ali J.A. Microbial Index and TNF-α, IL-4, CCL17 Level Among Burn Wound Infections in Hilla City, Iraq. Biomedical and Pharmacology Journal. 2016;9(3):1095-1100.
CrossRef - Al-Dahmoshi H.O.M. (2013). Genotypic and Phenotypic Investigation of Alginate Biofilm Formation among Pseudomonas Aeruginosa Isolated from Burn Victims in Babylon, Iraq. Science Journal of Microbiology. 2013.
- Al-Dahmoshi H.O.M. Genotypic Investigation of TypeIII Exotoxins Among Clinical and Environmental Pseudomonas spp. Isolates in Hilla City-Iraq. 2017.
- Finnan S, Morrissey J.P , O’gara F and Boyd E.F. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. Journal of clinical microbiology. 2004;42(12):5783-5792.
CrossRef - Mohammad H.H. Phenotypic Investigation for Virulence factors of Pyocine producing Pseudomonas aeruginosa Isolated from Burn Wounds, Iraq. International Journal of Scientific & Engineering Research. 2013;4(7):2114-21.
- Pirnay J.P, De Vos D, Duinslaeger L, Reper P, Vandenvelde C, Cornelis P and Vanderkelen A. Quantitation of Pseudomonas aeruginosa in wound biopsy samples: from bacte rial culture to rapid ‘real-time’polymerase chain reaction. Critical. 2000;4(4):255.
- Salman M, Ali A and Haque A. A novel multiplex PCR for detection of Pseudomonas aeruginosa: A major cause of wound infections. Pakistan journal of medical sciences. 2013;29(4):957.
CrossRef - Aghamollaei H, Moghaddam M.M, Kooshki H, Heiat M, Mirnejad R and Barzi N.S. Detection of Pseudomonas aeruginosa by a triplex polymerase chain reaction assay based on lasI/R and gyrB genes. Journal of infection and public health. 2015;8(4):314-322.
CrossRef - Rasheed M.N, Latteef N.S and Jassim A.B. Rapid Identification of Pseudomonas aeruginosa by Using Real Time PCR. Kufa. Journal For Veterinary Medical Sciences. 2017;7:(1B).
- Fadhil L, Al-Marzoqi A.H, Zahraa M.A and Shalan A.A. Molecular and Phenotypic Study of Virulence Genes in a Pathogenic Strain of Pseudomonas aeruginosa isolated from various clinical origins by PCR: Profiles of genes and Toxins. Research Journal of Pharmaceutical, Biological and Chemical Sciences. 2016;7(1):590-598.
- Gellatly S.L and Hancock R.E. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathogens and disease. 2013;67(3):159-173.
CrossRef - El-Din A.B, EL-Nagdy M.A, Badr R and EL-Sabagh A.M. Pseudomonas aeruginosa exotoxin A: its role in burn wound infection and wound healing. Egypt J Plast Reconstr Surg. 2008;32:59-65.
- Nikbin V.S, Aslani M.M, Sharafi Z, Hashemipour M, Shahcheraghi F and Ebrahimipour G.H. Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious origins. Iranian journal of microbiology. 2012;4(3):118–123.
- Smith R.S and Iglewski B.H. P. aeruginosa quorum-sensing systems and virulence. Current opinion in microbiology.2003;6(1):56-60.
CrossRef - Juhas M, Eberl L and Tümmler B. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environmental microbiology. (2005;7(4):459-471.
CrossRef - Cowell B.A, Twining S.S, Hobden J.A, Kwong M.S and Fleiszig S.M. Mutation of lasA and lasB reduces Pseudomonas aeruginosa invasion of epithelial cells. Microbiology. 2003;149(8):2291-2299.
CrossRef - Lomholt J.A, Poulsen K and Kilian M. Epidemic population structure of Pseudomonas aeruginosa: evidence for a clone that is pathogenic to the eye and that has a distinct combination of virulence factors. Infection and immunity. 2001;69(10):6284-6295.
CrossRef - Spilker T, Coenye T, Vandamme P and LiPuma J.J. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. Journal of clinical microbiology. (2004);42(5):2074-2079.
CrossRef - Khan A.A and Cerniglia C.E. Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of the exotoxin A gene using PCR. Applied and environmental microbiology. 1994;60(10):3739-3745.
- De Vos D.A. I.E.L., Lim A.N, Pirnay, J.P, Struelens M, Vandenvelde C, Duinslaeger L, Vanderkelen A and Cornelis P. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. Journal of clinical microbiology. 1997;35(6):1295-1299.
- Zhu H, Bandara R , Conibear T.C, Thuruthyil S.J, Rice S.A, Kjelleberg S, Givskov M and Willcox M.D. Pseudomonas aeruginosa with lasI quorum-sensing deficiency during corneal infection. Investigative ophthalmology & visual science. 2004;45(6):1897-1903.
CrossRef - Gholami A, Majidpour A , Talebi-Taher M, Boustanshenas M and Adabi M. PCR-based assay for the rapid and precise distinction of Pseudomonas aeruginosa from other Pseudomonas species recovered from burns patients. Journal of preventive medicine and hygiene. 2016;57(2):E81.
- Serra R, Grande R, Butrico L, Rossi A, Settimio U.F, Caroleo B, Amato B, Gallelli L and de Franciscis S. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert review of anti-infective therapy. 2015; 13(5):605-613.
CrossRef - Tumbarello M, De Pascale G, Trecarichi E.M, Spanu T, Antonicelli F, Maviglia R, Pennisi M.A, Bello G and Antonelli M. Clinical outcomes of Pseudomonas aeruginosa pneumonia in intensive care unit patients. Intensive care medicine. 2013;39(4):682-692.
CrossRef - Al-Wrafy F, Brzozowska E, Górska S and Gamian A. Pathogenic factors of Pseudomonas aeruginosa–the role of biofilm in pathogenicity and as a target for phage therapy. Postepy Hig Med Dosw (online). 2016;70:78-9.
- Chatterjee M, Anju C.P, Biswas L, Kumar V.A, Mohan C.G and Biswa, R. Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. International Journal of Medical Microbiology. 2016;306(1):48-58.
CrossRef - Fu W, Forster T., Mayer O, Curtin J.J, Lehman S.M and Donlan R.M. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrobial agents and chemotherapy. 2010;54(1):397-404.
CrossRef - Deep A, Chaudhary U and Gupta V. Quorum sensing and bacterial pathogenicity: from molecules to disease. Journal of laboratory physicians. 2011;3(1):4.
CrossRef - Moghaddam M.M, Khodi S and Mirhosseini A. Quorum Sensing in Bacteria and a Glance on Pseudomonas aeruginosa. Clinical Microbiology. 2014;3:156.
CrossRef - Wessel A.K, Liew J, Kwon T, Marcotte E.M and Whiteley M.. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. Journal of bacteriology. 2013;195(2):213-219.
CrossRef - Al-Khafaji N.S. Molecular Study of Some Virulence Factors among Pseudomonsa aeruginosa Recovered from Burn infection, Iraq. International Journal of Medicine and Pharmaceutical Sciences. 2014;4(3):71-80.
- Al-Alaq F.T Abdulazeem L, Al-Dahmoshi H.O, Al-Khafaji N.S, Al-Wesawei Y.A. PCR-based investigation of oxygenase among crude oil degrading bacteria in Hilla city, Iraq. International Journal of Pharm Tech Research. 2016;9(5):284-91.








