Sree Narayana Institute of Technology, Vadakkevila, Kollam, Kerala, 691 010, India.
Corresponding Author E-mail: sajeevjal@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1408
Abstract
This paper presents a tool to retrieve protein sequences along with features from Uniprot Database, given a set of Proteins IDs. These target IDs are stored in a spread sheet file. The tool uses AAIndex Database to retrieve 544 different features along with Amino acid count. The sequence and its features are displayed in a formatted table. This tool works with the help of Javascript, PHP, AJAX and PHP-Excel-API.
Keywords
Bioinformatics, Proteomics, Fasta, Protein, Uniprot, PHP, Ajax, LAMP
Download this article as:| Copy the following to cite this article: Sajeev J, Mahalakshmi T. WEBBIORET – A WebTool for Accessing Multiple Biological Sequences with Features. Biomed Pharmacol J 2018;11(1). |
| Copy the following to cite this URL: Sajeev J, Mahalakshmi T. WEBBIORET – A WebTool for Accessing Multiple Biological Sequences with Features. Biomed Pharmacol J 2018;11(1). Available from: http://biomedpharmajournal.org/?p=18854 |
Introduction
Bioinformatics is an interdisciplinary field which is mainly utilized to develop methods and software tools for understanding different types of biological data[1]. Biological data can be broadly classified as Genomics and Proteomics Data.
Drug discovery [2] is a crucial implementation area of Bioinformatics and ever going research is taking place in that domain for years. One such sub area is the studies relating to cellular activities and disease states in humans and other organisms[1]. Identification of DNA and protein sequences, protein domains and protein structures are very crucial at this point. So it can be understood that protein sequences are mandatory for these research activities.
The challenge faced by the researcher is to retrieve protein sequences in bulk numbers from well known databases such as NCBI, PDB, Uniprot, etc. In most of the cases the researcher need to search with the protein id and copy the sequence from the global databases.
Another issue is with the different formats of files given by the protein datasets. FASTA [3] is such a file type. It has a specific structure. The proposed tool bridges problems such as a) retrieve the sequence from the FASTA format and all other features such as sequence id, sequence description etc b) derive essential features according to the user attribute requirements c) give appropriate presentation of the data.
Background
In this section we discuss the important background information pertinent to this proposed work like Uniprot, AJAX and LAMP Server.
UniProt
The Universal Protein Resource (UniProt) is a commonly used data base for protein research. The data set is available in three categories such as UniProt Knowledgebase (UniProtKB), the UniProt Reference Clusters (UniRef), and the UniProt Archive (UniParc)[3] [4]. Uniprot’s development was closely tied up with TrEMBL and Swiss-Prot.
The reason for TrEMBL (Translated EMBL Nucleotide Sequence Data Library) development was due to the fact that the data was generated in a speed which couldn’t be managed by Swiss-Prot database alone. In 2002 the three institutes decided to combine their resources and expertise and formed the UniProt consortium[3] [4].
AJAX
Ajax expands to“Asynchronous JavaScript and XML”. This is a web technology. It is a group of interrelated programming techniques applied in the client browser side to create Web applications. With the help of Ajax, web programs can send transmit data without refresing page[6] .
LAMP Server
LAMP is an application server platform used to develop websites and web tools. The powerful PHP Web Application server in combination with the powerful Relational Database Management System makes a unique combination. Figure 1 clearly shows the different layers in the architecture. LAMP consists of Apache Webserver, MySql database and PHP Application server.
As a solution stack, LAMP is suitable for building interactive web softwares [7].
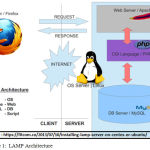 |
Figure 1: LAMP Architecture
|
AAIndex
AAindex is knows as a database of numerical values on behalf of different physicochemical attributes of amino acids [8]. AAindex comprises of three sections now:, AAindex1, AAindex2 and AAindex3 [8].
The complete database can be retrieved through the DBGET/LinkDB system at GenomeNet (http://www.genome.jp/dbget-bin/www b_nd?aaindex) [9] or downloaded by anonymous FTP (ftp://ftp.genome.jp/pub/db/community/aaindex/) [9]. Structures of Protein sequences and it’s functions are given by the combinations of physicochemical and biochemical attributes of 20 amino acids that are the building blocks of proteins[9]. Out of these properties an Amino acid Index can be created. From 222 amino acid indices Nakai et al. has done some research work to unearth the relationships among them using hierarchical cluster mechanisms [8][10]. Additionally, they released AAIndex2 after collection of 42 amino acid substitution matrices taken from literature.. Scientists are updating this AAIndex database in this manner [8] [12][13].
AAindex is in wide use especially in research of various protein analysis of organisms [8] , such as Protein subcellular localization prediction, hub protein prediction, membrane protein prediction etc[8]. AAindex has become a really notable resource in bioinformatics research[8]. The AAindex is released almost every year. The latest version which is available is the 9.0 release.[8].
The AAIndex1 currently contains 544 amino acid indices with its explanations[8]. For 20 amino acids each entry consists of a number called accession number, a short explanation of the index, the reference data and the numerical values for the protein properties[8] .
Proposed Tool
A web application is developed which takes an excel file as the maininput. This excel file contains the proteins ids where protein sequences are to be retrieved. The Figure 2 contains a few protein ids in a spread sheet.
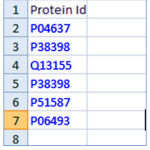 |
Figure 2: Sample data as protein ids in a spread sheet
|
Using the web application this file is selected and uploaded. Once the file is uploaded the web program will start reading the protein ids one by one using PHP excel API and start retrieving the sequence using UNIPROT web service along with the required feature set.
 |
Figure 3: Site Homepage
|
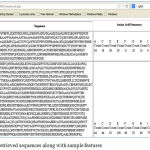 |
Figure 4: Retrieved sequences along with sample features
|
The web service retrieves the sequence in the form of FASTA file. The web application retrieves the sequence from the file and displays in the screen in a neat tabular format as given in the Figure 3. Here along with the sequence 20 amino acid frequencies are also extracted from the sequence as they are considered as important biomarkers which describe the physicochemical property of the protein[12].
Conclusion
The programmed tool retrieves any number of sequences with the help of the protein ids stored in the spreadsheet file. But the retrieved data is represented in the form of html table data along with feature set.
As a next step in this line the date retrieved will be stored in spreadsheet format along with the retrieved features. More than thousand amino acid features are relevant in the domain of proteomics research. All such features can be incorporated in the file and downloaded to the local file system of the researcher’s computer for further analysis and studies. Wavelet features are also in our list for the next version of our tool. The site can be viewed in the address http://www.snit.ac.in/research/.
Conflict of interest
There is no conflict of interest between authors.
References
- Opinion in Biotechnology, Volume 5, Issue 6, December 1994
- http://en.wikipedia.org/wiki/FASTA_format dated 18/4/2017 9.00 a.m.
- http://uniprot.org
- http://www.ebi.ac.uk/
- http://en.wikipedia.org/wiki/Ajax_(prorammin) dated 18/4/2017 9.00 a.m.
- http://en.wikipedia.org/wiki/LAMP_(28software_bundle)
- Kenta Nakai, Akinori Kidera, and Minoru Kanehisa. Cluster analysis of amino acid indices for prediction of protein structure and function. Protein Engineering, 2(2):93{100, 1988.
- http://www.nar.oxfordjournals.org
- Kawashima, P.Pokarowski,M.Pokaro S. Kawashima, P. Pokarowski, M. Pokarowska, A. Kolinski, T. Katayama, M. Kanehisa. “AAindex: amino acid index database, progress report 2008”, Nucleic Acids Research, 2007
CrossRef - Kentaro Tomii and Minoru Kanehisa. Analysis of amino acid indices and mutation matrices for sequence comparison and structure prediction of proteins. Protein Engineering, 9(1):27{36, 1996.
- Shuichi Kawashima and Minoru Kanehisa. Aaindex: amino acid index database. Nucleic acids research, 28(1):374{374, 2000.
- Shuichi Kawashima, Hiroyuki Ogata, and Minoru Kanehisa. Aaindex: amino acid index database. Nucleic Acids Research, 27(1):368{369, 1999.
- Esmaeil Ebrahimie, Mansour Ebrahimi, Mahdi Ebrahimi, “Amino acid features: a missing compartment of prediction of protein function”, Nature Proceedings, doi:10.1038./npre.2011.6693.1.








