Mary Shama, Kulandhaivel Murugesan and Hridhya Vijayan
Department of Microbiology, Karpagam University, Karpagam Academy of Higher Education, Coimbatore.
Corresponding Author E-mail: m.kulandhaiveljm@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1395
Abstract
Skin infections are very common throughout the world. Bacterial skin infections are the most common type of skin infection. The most common reported are impetigo, cellulitis, folliculitis, furunculosis, abscesses, scarlet fever, erysipelas, erythrasma, necrotizing fasciitis and some others. Specimens from 100 patients with different skin infection are collected aseptically with the aid of sterile swab. Bacterial species are isolated and identified by selective culture media and standard biochemical tests from the collected specimen. Out of 100 samples, 73 are found culture positive, gram negative isolates are predominant (89%), followed by gram positive isolates (10.9%). The most common isolates are Escherichia coli (57.5%), the predominant isolate, second most is Proteus sp. (31.5%) and the lowest percentage is recorded by Streptococcus pyogenes (10.9%). Among the 9 antibiotics, antibiotic sensitivity pattern of Cefaperazone/Sulbactum was found to be the most effective drug against the above two gram negative isolates and for the gram positive isolates – Penicillin and Ampicillin found to be most effective drugs.
Keywords
Antibiotic Susceptibility Testing Escherichia Coli; Proteus Sp., Skin Infection; Streptococcus Pyogenes;
Download this article as:| Copy the following to cite this article: Shama M, Murugesan K, Vijayan H. Isolation Identification and Antibiotic Sensitivity Pattern of Pyogens from Pyogenic Pathogens. Biomed Pharmacol J 2018;11(1). |
| Copy the following to cite this URL: Shama M, Murugesan K, Vijayan H. Isolation Identification and Antibiotic Sensitivity Pattern of Pyogens from Pyogenic Pathogens. Biomed Pharmacol J 2018;11(1). Available from: http://biomedpharmajournal.org/?p=18986 |
Introduction
Human skin acts as an excellent barrier to infection. Most bacteria live on our skin, in the nasopharynx, gastrointestinal tract and other parts of the body with little potential for causing disease because of the first line defense within the body. The surgical operation, trauma, burns, disease, nutrition and other factors affect the defenses. The skin barrier is disrupted by every skin incision and microbial contamination is inevitable, despite the best skin penetration.
Skin and skin structure infections are common and range from minor pyodermas to severe necrotizing infections. Skin can be infected by a variety of microorganisms ranging from bacteria to fungus and parasites. Bacterial skin infections are the most common. The most common gram positive organisms are hemolytic Streptococcus and Staphylococcus aureus. The gram negative rods include Pseudomonas aeruginosa, Escherichia coli, Enterobacter species, Klebsiella species and Proteus species (Efstrtiou., 1989).1 The fungal organisms are Candida species and moulds. There are many kinds of bacterial skin infections. The most common reported are impetigo, cellulitis, folliculitis, furunculosis, abscesses, scarlet fever, erysipelas, erythrasma, necrotizing fasciitis and some others.
The aim of this study is to determine the prevalence of bacterial pathogens associated with a skin infection and their drug sensitivity pattern.
Materials and Methods
Skin swabs were collected from a total of 100 patients with different kinds of skin infection. Samples were collected from patients in Medical College Hospital Trivandrum and KIMS Hospital Trivandrum, Kerala.
Culture Media Used
Blood agar,Mac Conkey’s agar, chocolate agar and Brain Heart Infusion Broth (BHI) for the bacterial isolation and identification. Muller – Hinton agar for Antimicrobial sensitivity testing.
Isolation and Identification of Bacterial Isolates
The swabs are streaked directly to the labeled agar plates and incubate 370C for 24 hr. The primary identification of the bacterial isolates was made based on the colony appearance and hemolysis. Identification and characterization of isolates were performed on the basis of colony characteristic, hemolysis, Gram staining and biochemical tests using standard microbiological methods. Biochemical tests applied were standard catalase, Indole production, Citrate Utilization Urease and Triple sugar iron. Biochemical characterization and identification of the bacterial isolates were done (Cowan and Steel, 1985)2
Antibiotics Susceptibility Testing
Antibiotic susceptibilities of bacterial isolates were determined according to the method recommended by the Clinical and Laboratory Standards Institute and Kirby Bauer Disc Diffusion method.3,4 The inoculumwas prepared for each bacterial isolate by adjusting the turbidity to 0.5 McFarland standard and spread on Muller-Hinton agar plates. Antibiotic discs (Himedia, Mumbai, India) were placed on the agar plates and incubated overnight at 37°C for 24 h. The zones of inhibition were measured in mm and the isolates were classified as sensitive, intermediate, and resistant according to CLSI tables and guidelines.5
Result
A total of 100 patient’s specimen was examined for different skin infections, 73 were found culture positive and 27 specimens were negative for growth. Out of which Gram Negative isolates were predominant (89%), followed by Gram Positive isolates. The most common isolates were Escherichia coli (57.5%), the predominant isolate, second most was Proteus sp. (31.5%) and the lowest percentage was recorded by Streptococcus pyogenes (10.9%) (Table 1, Fig 1). Gram negative bacteria were the dominant isolates (89%) from skin samples compared to Gram Positive bacteria. Antibiogram results from the present study show that Escherichia coli were more resistant to pencillin, cefotaxime while being least resistant to Cefaperazone/Sulbactum and gentamicin. Proteus sp. was more susceptible to tested antibiotics compared to Escherichia coli
Table 1: Bacterial Isolates from skin swab culture from different site collection
| SL.No | Isolates | Number | % |
| 1 | Escherichia coli | 42 | 57.5 |
| 2 | Proteus species | 23 | 31.5 |
| 3 | Streptococcus pyogenes | 8 | 10.9 |
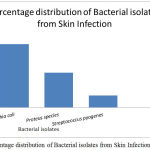 |
Figure 1: Percentage distribution of Bacterial isolates from Skin Infection.
|
Escherichia Coli
Escherichia coli is a Gram-negative, facultative, rod-shaped bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Good growth occurs in ordinary media. Colonies are large, thick, greyish white, moist, smooth opaque or partially translucent discs. On Mac Conkey medium colonies are bright pink due to lactose fermentation. It ferments all the sugars and produes acid and gas. Four main types of clinical syndromes are caused by E. coli – Pyogenic infection, Urinary tract infection, diarrhea and gastroenteritis.
Proteus Species
Proteus bacilli are widely distributed in nature as saprophytes, being found in decomposing animal matter, sewage, manure soil, and human and animal feces. They are opportunistic pathogens, commonly responsible for wound infections, bronchopneumonia, cystitis and urolithiasis, septicemia. It is Gram negative rod, motile, non-spore forming, non-encapsulated, facultative anaerobic. Cultures of Proteus bacilli have a characteristic putrefactive odour – Fishy odour. Swarming growth occur on solid culture media. Swarming does not occur on Mac Conkey medium, on which the smooth colourless colonies are formed. Proteus species do not usually ferment lactose, but have shown to be capable lactose fermenters depending on the species in a triple sugar iron (TSI) test. It is oxidase negative but catalase and nitrate positive. It has the ability to degrade the urea to ammonia, by the production of the enzyme urease.
Streptococcus pyogenes or Group A Streptococcus.
S. pyogenesis the cause of many important human diseases, ranging from mild superficial skin infections to life-threatening systemic diseases. S.pyogenes is the gram positive cocci arranged in chains or pairs. It is an aerobe and facultative anaerobe. It is exacting in nutritive requirement, growth occurs in media containing fermentable carbohydrates or enriched with blood or serum. On blood agar, the colonies are small, circular, semitransparent, low convex discs with an area of clear hemolysis around them. Streptococcus pyogenes ferment lactose, sucrose, mannitol, glucose and produce acid.
Antibiotic Sensitivity Pattern of Bacterial Isolates
The commonest bacterial pathogen isolated from pyogenic infections followed by E.coli (57.5%), Proteus species (31.5%) and Streptococcus pyogenes (10.9%). The percentage of bacterial isolates towards Penicillin, Ampcillin, Cotrimoxazole, Cefaperazone, Cefotaxime, Netilmicin, Levofloxacin, Ofloxacin and Gentamicin were tabulated in Table 2 and Fig 2. The Gram negative pathogen, E.coli shown maximum resistance towards Penicillin (95.25%), Ampcillin (85.71%), Cotrimoxazole (76.19%), Cefotaxime (80.95%), Netilmicin (61.9%) and Levofloxacin (57.14 %) where as Proteus species have maximum resistance towards Cotrimoxazole (76.25%). In case of gram positive bacteria, Streptococcus pyogenes were resistance towards Cotrimoxazole (62.5%), Cefotaxime (75%), Netilmicin (62.5%) and Gentamicin (87.5%).
Table 2: Antibiotic Sensitivity Pattern of Bacterial Isolates.
| Antibiotics | Unit | Escherichia coli | Proteus species | Streptococcus pyogenes | |||
| %S | %R | %S | %R | %S | %R | ||
| Penicillin | 1 Unit | 4.76 | 95.2 | 56.52 | 4.3 | 100 | 0 |
| Ampcillin | 10 mcg | 9.52 | 85.71 | 69.56 | 30.4 | 100 | 0 |
| Cotrimoxazole | 25 mcg | 23.80 | 76.19 | 21.73 | 78.26 | 37.5 | 62.5 |
| Cefaperazone/Sulbactum | 30 mcg | 90.47 | 9.5 | 95.6 | 4.3 | – | – |
| Cefotaxime | 30 mcg | 19.04 | 80.95 | 69.56 | 30.4 | 25 | 75 |
| Netilmicin | 30 mcg | 38.0 | 61.9 | 60.8 | 39.13 | 37.5 | 62.5 |
| Levofloxacin | 5 mcg | 42.85 | 57.14 | 82.60 | 17.39 | 50 | 50 |
| Ofloxacin | 5 mcg | 52.3 | 47.6 | 65.21 | 34.78 | 50 | 50 |
| Gentamicin | 10 mcg | 71.42 | 28.5 | 78.26 | 21.73 | 12.5 | 87. |
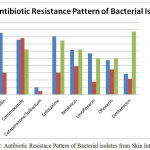 |
Figure 2: Antibiotic Resistace Pattern of Bacterial isolates from Skin Infection.
|
Discussion
Pyogenic infection is referred to bacterial infection that leads to severe local inflammation with pus. The invasion and multiplication pathogens in tissue will cause cell damage and leads to loss of integrity of tissue and skin. This will leads to subcutaneous infection to life threatening complications. The present study is aimed to isolate the bacterial pathogens which cause pyogenic infection and study their antibiotic resistance pattern. In this study, both gram positive and gram negative pathogens were isolated from a total of 100 samples. The predominant pathogens were gram negative bacteria. It was agreed with a previous studies Ghosh et al6 and Zubair et al,7 in their studies the aerobics growth of pus culture the dominance pathogens were Gram negative bacterias. E.coli (57.7%), one of the most commonest and predominant pathogen and Proteus species (31.5%) and followed by gram positive pathogens Streptococcus pyogenes (10.9%). A previous report states that out of 59.3% of gram negative bacteria, the predominant pathogen was E.coli (21.7%), Klebsiella (16.8%), Pseudomonas aeruginosa (7.5%), Proteus species (7.1%) and Acinetobacter species (6.7%) where as gram positive bacteria (40.7%) like Staphylococcus aureus (37.2%), Coagulase negative Staphylococcus aureus (1.3%) and Streptococcus pyogenes (2.2%) (Mantravadi et al.,2015).8 From another report it was found that E.coli (Basu et al., 2009)9 and Pseudomonas (Raza et al., 2013)10 were the most predominant gram negative pathogen occur in wound infections. The antibiotic resistance pathogens were rapidly increased due to the frequent use of antibiotics. Now a day it became great difficulty to manage or control the pyogenic pathogen and one of the major problems faced by the physicians (Singh et al., 2013).11 In this study the gram positive pathogen, Streptococcus pyogenes shows resistance towards Gentamicin (87.5%), Netilmicin (62.5%), Cotrimoxazole (62.5%) and sensitive to Penicillin (100%) and Ampicillin (100%) and intermediate towards Levofloxacin (50%) and oflaxacin (50%). These findings were similar to those of Manthravadi et al,8 and Rao et al.12 In other hand, most of the gram negative pathogens were highly resistance towards Sulfamethoxazole, Cephalosporin, Fluroquinolones and sensitive to aminoglycosides. These findings were agreed with the previous studies (Mantravadi et al., 2015).8 The combination of antibiotics Cefaperazone + Sulbactum shows maximum sensitivity of about 90-95%. It was correlated with the previous studies done by Javeed et al.13 Rao et al,12and Anguzu and Olila.14
Conclusion
This study revealed the presence of skin infection caused by bacteria, those were capable of causing various human illness. The bacterial isolates screened in various skin infections were Escherichia coli (57.5%), Proteus species (31.5%), Streptococcus pyogenes (10.9%). The bacterial isolates from the skin infection in this study predominately were Escherichia coli, compared with to others. Bacterial isolates exhibited high to moderate levels of resistance against different classes of antibiotics.
References
- Efstrtiou Androulla: Outbreaks of human infection caused by pyogenic Streptococci of Lancefield groups C and G. Journal of Medical Microbiology. 1989;29:207-219.
CrossRef - Cowan S.T, Steel K.J. Manual for the identification of medical bacterial. 2nd Cambridge University Press London. 1993.
- National Committee for Clinical Laboratory Standards, Methods for Disk Susceptibility Tests for Bacteria That Grow Aerobically, NCCLS Document M2-A7, Wayne, National Committee for Clinical Laboratory Standards 7th edition. 2000.
- Bauer W.A, Kirby M.W, Sherris J.C, Truck M.M. Antibiotic Susceptibility testing by standardized single disc method Am. J Clin. Pathol. 1966.
- Performance standards for antimicrobial susceptibility testing. Twentieth informational supplement, Clinical and Laboratory Standards Institute Doc. M100eS20. 2010.
- Ghosh A, Karmakar P.S, Pal J, Chakraborty N, Debnath N.B, Mukherjee J.D. Bacterial incidence and antibiotic sensitivity pattern in moderate and severe infections in hospitalized patients. J Indian Med Assoc. 2009;107(1):21–2, 24–5.
- Zubair M, Malik A, Ahmad J. Clinico-microbiological study and antimicrobial drug resistance profile of diabetic foot infections in north India. Foot (Edinb). 2011;21(1):6–14.
CrossRef - Bindu H.M, Reddy M.C, Shravani V. Aerobic isolates in pus and their antibiotic sensitivity pattern: a study conducted in a teaching hospital in Andhra Pradesh. International Journal of Medical Science and Public Health. 2015;4(8):1076-1079.
CrossRef - Basu S, Panray R.T, Singh B.T, Gulati A.K, Shukla V.K. A prospective, descriptive study to identify the microbiological profile of chronic wounds in outpatients. Ostomy Wound Manage. 2009;55(1):14–20.
- Raza M.S, Chander A, Ranabhat A. Antimicrobial susceptibility patterns of bacterial isolates in postoperative wound infections in a tertiary care hospital, Kathmandu, Nepal. Open J Med Microbiol. 2013;3:159–63.
CrossRef - Singh S, Khare M, Kumar R.P, Bagde S, Sahare K.N, Dwivedi D and Singh V. Antibacterial Activities Against Pyogenic Pathogens. International Journal of Pharmaceutical Sciences and Research. 2013;4(8):2974-2979.
- DVMVSVR R, Basu R, Biswas D.B. Aerobic bacterial profile and antimicrobial susceptibility pattern of pus isolates in a south Indian tertiary care hospital. IOSR J Dent Med Sci. 2014;13(3):59–62.
CrossRef - Javeed I, Hafeez R, Anwar M.S. Antibiotic susceptibility pattern of bacterial isolates from patients admitted to a tertiary care hospital in Lahore. Biomedica. 2011;27:19–23.
- Anguzu J.R, Olila D. Drug sensitivity patterns of bacterial isolates from septic post-operative wounds in a regional referral hospital in Uganda.2007;7(3):148–54.








