Priya Durairaj , Rohithkumar Reddy, Thirunavukkarasu Palaniyandi, Rajeswari Hari and Saravanan T. S.
, Rohithkumar Reddy, Thirunavukkarasu Palaniyandi, Rajeswari Hari and Saravanan T. S.
Department of Biotechnology, Dr. M.G.R Educational and Research Institute University, Maduravoyal, Chennai-95, India.
Corresponding Author E-mail: priyadurairajvit@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1379
Abstract
To evaluate the effect of mutation in FMR1 protein on the binding energy of protein interaction by homology modeling and docking study using Bioinformatics approach. As per NCBI/Swiss Prot database information normal and mutated FMR1 proteins with accession number AAB28395 and rs121434622, respectively and interacting proteins Cytoplasmic FMR-1 interacting protein, Cytoplasmic FMR1 interacting protein 2, Pre mRNA 3’- end- processing, Tudor domain-containing protein3 (TDRD3), Kinesin like protein KIF3C (KIF3C) and Microspherule protein 1 (MCRS1) has been modeled by using the web server MODWEB for tertiary structures and thereafter every interacting protein was allowed to dock with normal and mutated FMR1 protein by involving HEX server to record the changes in binding energy resultant of mutation. Based on the homology modeling approach, tertiary structures of all studied proteins were successfully modeled and further in docking analysis it has been observed that mutated FMR1 protein highlighted decreased change in the binding energy as compared to normal FMR1 protein docking. Based on the bioinformatics approaches, our study confirms that lowering change in the binding energy for the interacting protein with mutated FMR1 protein when compared to normal FMR1 protein does clearly affect the protein - protein interaction and hence it lead to Fragile X syndrome in affected patients showcasing such mutations.
Keywords
Binding Energy; FMR1; Fragile X Syndrome; Protein Docking; Protein Interaction
Download this article as:| Copy the following to cite this article: Durairaj P, Reddy R, Palaniyandi T, Hari R, Saravanan T. S. Insilico Analysis of Fmrp Protein in Fragile X Syndrome. Biomed Pharmacol J 2018;11(1). |
| Copy the following to cite this URL: Durairaj P, Reddy R, Palaniyandi T, Hari R, Saravanan T. S. Insilico Analysis of Fmrp Protein in Fragile X Syndrome. Biomed Pharmacol J 2018;11(1). Available from: http://biomedpharmajournal.org/?p=19620 |
Introduction
Fragile X syndrome (FXS) is one of the most common inherited forms of intellectual disability (ID).1,2 It happens because of expansion of CGG repeat sequence in the first exon of the FMR1 gene, gives rise to transcriptional silencing of the gene and resultant low or absence of gene product, fragile X mental retardation protein (FMRP).3 The protein involves in proper synaptic plasticity, neuronal morphology, and cognitive development and down regulation leads to different levels of ID.4 Along with ID, other prominent phenotypic characteristics of FXS are narrow face, prominent forehead, protruding ears, high arched palate, strabismus, macro-orchidism and connective tissue dysplasia.2,5 Males are more severely affected than females as the gene localization remained on X chromosome and due to the presence of unaffected second X chromosome in female6
By using Bioinformatics approach, model experimentally determined protein structures are primarily used in the docking and interaction studies. These stable protein structures could be used by involving docking specific algorithms those have been developed further to understand the protein-protein/protein-ligand interaction. The software can perform protein docking which is the task to calculate the 3D structure of a protein complex from its unbound or model- built subunits. Many protein docking assume proteins are rigid and involves geometric7 or fast Fourier transform (FFT) correlation techniques8 to find a relatively small number of putative docking orientations which could be improved by using sophisticated techniques. To name several protein docking programs those have been made available as web servers are RosettaDOck,9,10 HadDock11 and PatchDocksever.12 All these servers provide detailed structural interaction with statistically significant algorithms. In the present study, FMRP protein which brings about the FXS in human has been investigated for their reported mutation and resultant binding energy changes occurring with selected interacting proteins has been reported by involving Bioinformatics modeling and docking approach.
Material and Methods
Retrieval of Fragile X mental retardation protein (FMRP) and other interacting proteins sequences
In the present study to confirm the structural changes brought by mutation/s in the FMRP protein and resultant binding energy variations with the interacting proteins has been carried out by using the modeling and docking approach.
In requirement, reported normal FMRP (n- normal) and mutated human FMRP (m-mutated) proteins sequences were retrieved from NCBI protein database with accession number AAB28395 12 and rs12143462213 respectively, where later one entry having amino acid transition as ILE304ASN as available in the SNPs database. Likewise, many interacting proteins whose structural activity relationship needs to be investigated were also retrieved for the protein sequences from Swiss –Prot database with the codes as given below:
Cytoplasmic FMR1 interacting protein 1 (CYFIP1)code : Q7L576 14
Cytoplasmic FMR1 interacting protein 2 (CYFIP 2) code: Q96F07 14
Pre mRNA 3’- end- processing factor (FIP1) code: Q6UN1515
Tudor domain-containing protein3 (TDRD3) code: Q9H7E26
Kinesin like protein KIF3C (KIF3C) code: O1478216
Microspherule protein 1 (MCRS1) code: Q96EZ817
BLASTP analysis and Template Search
In a next step to determine best scored structural homolog as a template, all normal and mutated FMRP protein as well as other six interacting proteins was checked for the sequence homology by searching against sequence datasets of which structures were determined by X-ray crystallography and record available as PDB ID code. BLASTP program searched through database and suitable template with its PDB ID was retrieved further to build the model tertiary structures for the input sequences by using modeling server. BLASTP is available at: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins.
Protein Modeling (Homology Modeling)
Best scored template confirmed in the last step was used further to build the tertiary structure model of the query sequence with the involving web server ‘MODBASE’ available at https://modbase.compbio.ucsf.edu/modweb/.18 Server accepts the template and input sequence and by involving the modeler algorithm, built up the 3D structure which was downloaded in PDB format and further checked for its quality by Ramachandran plot.
Ramachandran Plot Analysis
Modeled structures were checked for its prediction quality as per Ramachandran plot rules available at www.biochem.ucl.ac.uk/raman/procheck/procheck.19 Server checked for stereo-chemical quality of structure and reported amino acids placed in the allowed and disallowed regions.
Protein – Protein Docking
The probable structural changes occurred in the mutated FMRP (m) protein as compared to the normal FMRP (n) was estimated when the binding energy of every interacting protein with normal and mutated FMRP was compared in Hex protein interaction server available at http://hex.loria.fr/.20 Any deviation from the native structure should affect protein-protein binding energy which may either increase or decrease with type of mutation persists. These variations in the binding energy in normal versus mutated protein groups are considered as a base for the recurring metabolic disorders in affected patients brought about by mutation in FMRP (m).
Result
Template Search by BLASTP
As per BLASTP with specific structural homolog search, for both normal and mutated FMRP protein, common template was identified as PDBID: 2BKD_ N encoding the structure of N-terminal domain of Fragile X mental retardation protein (Homo sapiens). Hence, for normal FMRP (n) protein, three dimensional structure with PDBID code 2BKD_ N was taken as standard and downloaded and thereafter used in docking analysis with other interacting protein. On the other hand, mutated FMRP (m) protein was modeled as predicted structure by using 2KBD as a template so that any change in the amino acid position brought by mutation could be detected as altered structure.
In interacting protein set, CYF1P1 and CYF2P2 protein found common structural homolog as PDBID 3P8C_A encoding Cytoplasmic FMR1-interacting protein 1 of H. sapiens; FIP protein showed homology with Uricase enzyme having PDB ID code 2YZB expressing in Arthrobacter globiformis; TDRD3 protein found template homolog with PDB ID 3PNW encoding Tudor domain-containing protein 3 of Homo sapiens; 5KIF3C protein showed structural homolog with PDBID 2B6V which is the theoretical model encoding P2Y14 receptor of H. sapiens; MSP58 protein showed sequence homology with homolog with PDB ID 2X5F encoding Aspartate_tyrosine_phenylalanine pyridoxal-5′ phosphate-dependent amino transferase expressed by Staphylococcus aureus. The other details have been showcased in Table 1 such as score, E- value and related data.
Table 1: BLASTP based RSCB PDB database specific search recorded template homolog for every query protein sequence
| S.No. | Protein Identifier | PDB ID | Query sequence length | Query sequence coverage | Bit scores | Percentage of identity | E- Value |
| 1 | FMRP(n)/(m) | 2BKD_ N | 632 | 21% | 280 | 131/134 | 0.87 |
| 2 | CYF1P1 | 3P8C_A | 1253 | 100% | 2616 | 1253/1253 | 0 |
| 3 | CYF2P2 | 3P8C_A | 1278 | 99% | 2311 | 1098/1278 | 0 |
| 4 | FIP | 2YZB | 594 | 7% | 34.7 | 37/142 | 0.12 |
| 5 | TDRD3 | 3PNW | 653 | 11% | 165 | 76/76 | 0.0005 |
| 6 | 5KIF3C | 2B6V | 793 | 47% | 782 | 376/377 | 0 |
| 7 | MSP58 | 2X5F | 462 | 13% | 29.3 | 17/66 | 4.1 |
Protein Modeling
By considering aforementioned best scored template as a standard and with input protein sequences; server MODWEB modeled following structures successfully as given below:
 |
Figure 1: MODWEB server based modeled structures for proteins
|
Structure Quality Check
As per Ramachandran plot analysis more than 90% amino acids of modeled proteins showcased the amino acid positioning in favored region except for the protein FIP where 77.6% amino acids are positioned in favored region. Overall, it has been concluded that structures are validated in interaction studies as per plot analysis (Table 2).
Table 2: Ramachandran Plot score values represented for each modeled protein
| Name of the protein | Favoured regions | Allowed regions | Disallowed regions | Generously allowed regions | Total no. of residues | G factor | Protein quality | E(KJ/mole) |
| FMRP (m) | 90.2 | 16.2 | 6.3 | 6.3 | 131 | 0.7 | Better | -76146 |
| CYF1P1 | 94.5 | 5.3 | 0 | 0.2 | 2685 | 0.6 | Good | -8601.54 |
| CYF1P2 | 93.4 | 5.4 | 0.7 | 0.4 | 1270 | 0.6 | Good | -8601.44 |
| TDRD3 | 92.5 | 7.5 | 0 | 0 | 689 | 0.6 | Good | -1331.9 |
| 5KIF3C | 92.1 | 6.5 | 0.3 | 0.3 | 456 | 0.6 | Good | -4318.52 |
| MSP58 | 92.7 | 6.3 | 0.6 | 0.3 | 374 | 0.6 | Good | -22794.1 |
| FIP | 77.6 | 17.6 | 3.2 | 1.6 | 153 | 0.6 | Better | -6801.24 |
Protein-Protein Docking study
After modeling of the mutated FMRP protein along with other interacting proteins, all are subjected to the docking analysis. In the process, six interacting proteins when allowed to dock with FMRP (n) and FMRP (m) it has been observed that binding energy results from the normal and mutant FMRP protein remained varied with common interacting protein. For ex: normal FMRP interacted with CYF1P1 with energy -8.858359 KJ/mol where in case of mutant FMRP it was recorded on lower side as -9.554142 KJ/mol. This variation in the binding energy clearly indicated structural variation occurring due to the mutation and that also lead to change in interaction of involving proteins. As all proteins showcased defined variation in the binding energy with mutated FMRP protein compared to normal protein that clearly directed us, any mutation which leads to amino acid transition certainly can halt functional process of protein interaction and linked metabolism (Table 3 and Fig. 2).
Table 3: Variation in binding energy recorded in mutated FMRP proteins with interacting proteins as compared to normal FMRP.
| Interacting proteins | Normal FMRP | Mutated FMRP |
| Binding energy (KJ/mol) | ||
| CYF1P1 | -8.858359 | -9.554142 |
| CYF1P2 | -8.34569 | -9.44426 |
| TDRD3 | -6.53478 | -7.526271 |
| MSP58 | -5.982772 | -7.732903 |
| 5KIF3C | -6.514271 | -9.554142 |
| FIP | -5.782663 | -8.228176 |
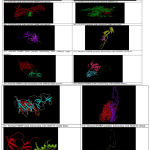 |
Figure 2: Deviation in the protein- protein interaction was observed and related with changes in binding energy.
|
Discussion
In the present study it has been clearly evidenced based on Bioinformatics approach which involves the sequence and structural homology approach along with protein-protein docking that any specific mutation in FMRP with resultant amino acid transition produces defined protein structural changes. Here it has been related that structural changes via mutation in protein leads to associated binding energy variation when other protein interacts with them.
In the present study it has been evidenced that interacting proteins named as CYFP1, CYFP2, TDRD3, MSP58, 5KIF3C and FIP profoundly showcased variation in binding energy when docked with normal and mutated FMRP proteins. This has indicated that SNPs based amino acid transition in mutated proteins lead to changes in the structure and function and probably this is the reason that it gives diseased condition in patients as evidenced by particular mutation detected such as ILE304ASN transition in FMRP.
As per earlier reports mutation in the FMR1 gene which leads to Fragile X mental retardation syndrome such as Ile 304-to-Asn (I304N) substitution reported by 21 also transition as (367N) was reported by.22 Not only the positional mutation but also frame shift mutation and premature termination of protein in exon 5 of the FMR1 gene due to 1 Bp deletion (373 del A) has also been reported. This situation leads to complete loss of FMRP protein as evidenced by western blot and related directly with fragile X syndrome.23 Further23 reported mutation in the adult male with classic fragile X syndrome was identified as 2 base pairs change (23714GG-TA) of FMR1 gene which resulted in 2 products as per RT-PCR and sequence analysis but by western blot they did not find any protein expressed and that leads to fragile X syndrome in affected person.
Another worker24 noted that I304N mutation on second KH domain of FMR1 is vital in stabilizing sequence-specific RNA-protein interactions. He reported that this mutation abrogates the interaction of the FMR1 KH2 domain with its target, kissing complex RNA. In our study also similar feature was recorded where mutation brought about the changes in the interaction of most proteins with FMRP. In accordance to our study,25 reported that I304N substitution profoundly reduced FMR1 homooligomerization and abrogated the pairing between FMR1 oligomer and stress granule protein TDRD3.
It has been observed that the mutation in coding region of FMRP leads to changes on the structural level as evidenced in the study and that gives variation in binding affinity of interacting proteins with FMRP and this ultimately affects the metabolome which leads to Fragile X mental retardation syndrome in patients as confirmed by the Bioinformatics approach.
Conclusion
Study successfully highlighted the importance of Bioinformatics approach to assess the impact of mutation on the structural level of FMRP protein. Resultant structural rearrangement of amino acids of FMRP protein certainly interacted in different ways as compared to normal protein’s amino acids and resultant variation in the binding energy with interacting proteins was evidenced as compared to normal protein interaction. Study by involving the bioinformatics’ tools such as alignment, modeling, QSAR and protein-protein interaction demonstrated that the PMRP protein mutation leads to structural changes and when they interact with the other proteins defined change rather increasing, increase in binding energy was observed which further demonstrate that probably it will change the functionality of the proteins. Such study demonstrated the exact molecular output of the mutation that ultimately represents the picture of the syndrome in the sufferers.
Acknowledgement
We thank Dr.M.G.R Educational and Research Institute University and Vellore Institute of Technology for providing the necessary facilities.
Conflict of Interest
There is no conflict of interest
References
- Martyn M., Anderson V., Archibald A. Offering fragile X syndrome carrier screening: a prospective mixed-methods observational study comparing carrier screening of pregnant and non-pregnant women in the general population. BMJ. 2013;9:e00366. 0pmid: 24022395.
- Hersh J. H., Saul R. A. Committee on Genetics. Health supervision for children with fragile X syndrome. Pediatrics. 2011;127(5):994–1006.
CrossRef - Santoro M. R., Bray S. M., Warren S. T. Molecular mechanisms of fragile X syndrome a twenty-year perspective. Annu Rev Pathol. 2012;7:219–245.
CrossRef - McLennan Y., Polussa J., Tassone F., Hagerman R. Fragile X syndrome. Curr Genomics. 2011;12(3):216–224.
CrossRef - Wang J., Huo K., Ma L., Tang L., Li D., Huang X., Yuan Y., Li C., Wang W., Guan W., Chen H., Jin C, Wei J., Zhang W., Yang Y., Liu Q., Zhou Y., Zhang C., Wu Z., Xu W., Zhang Y.,Liu T., Yu D., Zhang Y., Chen L., Zhu D., Zhong X., Kang L., Gan X., Yu X., Ma Q., Yan J., Zhou L., Liu Z., Zhu Y., Zhou T., He F., Yang X. Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol. 2015;75-36.
- David W. R.,Venkatraman V. Ultra-fast FFT protein docking on graphics processors. 2010;26:2398-2405.
- Ui-Hang E. S., Xiang-Ru M., Li-Li S . C., Choon-Weng L., Narayanan K. Predicted interaction of human Ribosomal Protein S15 with Fragile X Mental Retardation Protein. Journal of Applied Biology & Biotechnology. 2016;4(02):038-045.
- Lyskov S., Gray J. J. The Rosetta Dock server for local protein-protein docking. Nucleic Acids Res. 2012;1:233-238.
- Dominguez C., Boelens R., Bonvin A. M. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2013;125(7):1731-1737.
CrossRef - Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2015;1:363-367.
- Verkerk A. J., de Graaff E., Boulle K. D., Eichler E. E., Konecki D. S., Reyniers E., Manca A., Poustka A., Willems P. J., Nelson D. L. Alternative splicing in the fragile X gene FMR1. Hum Mol Genet. 2013;2(8):1348.
CrossRef - Ross M. T. The DNA sequence of the human X chromosome. Nature. 2015;434(7031):325-327.
CrossRef - Nebel R. A., Zhao D., Pedrosa E., Kirschen J., Lachman H. M., Zheng D., Abrahams B. S. Reduced CYFIP1 in Human Neural Progenitors Results in Dysregulation of Schizophrenia and Epilepsy Gene Networks. 2016;11(1):e0148039.
- Alrwas A., Quesada J. R., Marcos L. A., Mehta S. S., Shattuck B. L., Nguyen N. D., Juneja H. S. Case of polycythemia vera concurrent with FIP1L1-PDGFRA-positive myeloproliferative neoplasm with eosinophilia. 2012;205(10):519-22. doi: 10.1016/j.cancergen.2012.05.010.
CrossRef - Pichlmair A., Kandasamy K., Alvisi G., Mulhern O., Sacco R., Habjan M., Binder M., Stefanovic A., Eberle C. A., Goncalves A., Bürckstümmer T., Müller A. C., Fauster A., Holze C., Lindsten K., Goodbourn S., Kochs G., Weber F., Bartenschlager R., Bowie A. G., Bennett K. .L, Colinge J., Superti-Furga G. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487(7408):486-90.
CrossRef - Davidovic L., Bechara E., Gravel M., Jaglin X. H., Tremblay S., Sik A., Bardoni B., Khandjian E. W. The nuclear microspherule protein 58 is a novel RNA-binding protein that interacts with fragile X mental retardation protein in polyribosomal mRNPs from neurons. Hum Mol Genet. 2016;15(9):1525-38.
CrossRef - Eswar N., John B., Mirkovic N., Fiser A., Ilyin V. A., Pieper U., Stuart A. C., Marti-Renom M. A., Madhusudhan M. S., Yerkovich B., Sali A. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2013;31(13):3375-80.
CrossRef - Vyas K., Ukawala R. D., Ghate M and Chintha C. Homology Modeling a Fast Tool for Drug Discovery: Current Perspectives. Indian J Pharm Sci. 2012;74(1):1–17.doi: 10.4103/0250-474 X.102537.
- Macindoe G., Mavridis L., Venkatraman V., Marie-Dominique D., David W. R. Hex Server an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2012;38:445-449.
CrossRef - Siomi H., Choi M., Siomi M. C., Nussbaum R. L., Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 2012;77:33-39.
CrossRef - Boulle K. D., Verkerk A. J., Reyniers M. H., Vits E., Hendrickx L., Van J., Roy B.,Den V. B., de Graaff F. E., Oostra B. A., Willems P. J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nature Genet. 2013;3:31-35.
CrossRef - Lugenbeel K. A., Peier A. M., Carson N. L., Chudley A. E., Nelson D. L. Intragenic loss of function mutations demonstrate the primary role of FMR1 in fragile X syndrome. Nature Genet. 2015;10:483-485.
CrossRef - Darnell J. C., Fraser C. E., Mostovetsky O., Stefani G., Jones T. A., Eddy S. R., Darnell R. B. Kissing complex RNAs mediate interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2015;19:903-918.
CrossRef - Linder B., Plottner O., Kroiss M., Hartmann E., Laggerbauer B., Meister G., Keidel E., Fischer U. Tdrd3 is a novel stress granule-associated protein interacting with the fragile-X syndrome protein FMRP. Hum Molec. Genet. 2008;17:3236-3246.
CrossRef








