Divya Sharma1, Surya Prasath V. B2,3,4, Vijayarajan V5, Mohan Kubendiran6 and Padmapriya R7
1,5,6,7SCOPE (School of Computing Science and Engineering), VIT University, Vellore, India.
2Computational Imaging and Vis Analysis (CIVA) Lab, Department of Computer Science, University of Missouri-Columbia, Columbia MO 65211 USA.
3Biomedical Informatics, Cincinnati Children's Hospital Medical Center (CCHMC), Cincinnati, OH 45229 USA.
4Department of Biomedical Informatics, College of Medicine, University of Cincinnati, OH USA.
Corresponding Author E-mail: vijayarajan.v@vit.ac.in
DOI : https://dx.doi.org/10.13005/bpj/1338
Abstract
Clustering of clinical documents is a major research area in the field of machine learning and artificial intelligence, which aims to acquaint some type of association with the information that helps to highlight relevant examples and patterns. The rich corpus of clinical notes consists of several unprocessed data that needs to be mined with appropriate techniques to improve and augment the existing healthcare system. Biomedical information mining is a general research strategy that aims to recover, break down, and analyze clinical information from a collection of medical and/or medicinal records. This paper presents a novel approach that utilizes Non-Negative Matrix Factorization (NMF) clustering approach to mine the medication names based on age of the patients. Pharmaceutical data from clinical notes is regularly communicated with prescription names and other medication information, which needs to be mined, based on the similarity between documents so that more accurate extraction of similarity could be accomplished. Even in the wake of being an exceptionally effective solution, clustering is yet not deployed in major search engines. The basic issue with it is to determine a fast and accurate cluster values even after reducing the complexity of the technique. This paper presents an enhanced multi-viewpoint similarity measure that utilizes many distinct viewpoints to measure similarity between documents so that more accurate extraction of similarity could be accomplished.
Keywords
Clinical Documents; Clustering Algorithms; Cluster Analysis; Heuristic; Text Mining; Semantics
Download this article as:| Copy the following to cite this article: Sharma D, Prasath S. V. B, Vijayarajan V, Kubendiran M, Padmapriya R. Enhanced Multi-View Point Non-Negative Matrix Factorization Clustering for Clinical Documents Analysis. Biomed Pharmacol J 2017;10(4). |
| Copy the following to cite this URL: Sharma D, Prasath S. V. B, Vijayarajan V, Kubendiran M, Padmapriya R. Enhanced Multi-View Point Non-Negative Matrix Factorization Clustering for Clinical Documents Analysis. Biomed Pharmacol J 2017;10(4). Available from: http://biomedpharmajournal.org/?p=18318 |
Introduction
Information mining is known to be a notable wellspring of knowledge retrieval techniques from database. It is considered as a counterfeit strategy that allows us to find helpful information dwelling in important parts of data sources. It has been demonstrated to have good potential to separate valuable data from a comprehensive gathering of information. Clustering has been one of the most proficient approaches that can provide the researchers a way to extract information from the grouped clinical notes. Various researches have been made in this domain that have doubtlessly accomplished incredible pace in the locale of restorative exploration and clinical practice. Clinical data mining is the way toward applying the information mining strategies on the acquired literary clinical reports.
Rich content information of clinical reports contains data about drugs and syndromes. Extricating this data has proven useful to refine the medical system. Many studies have proposed various proficient techniques for diagnosing diseases to extricate the right medical information from the unprocessed data. Clinical documents are broadly utilized for future investigation and determination of the sickness. The clinical notes have an incredible use in drug store so to lessen imitation and avoid drug misuse. Record clustering helps gather them into pertinent groups. This is done basically to find critical patterns to put a collection of the comparable articles into groups. By utilizing this method, we can perform grouping on literary information corpus that is in unstructured and semi-organized configuration. These sorts of important data separated from biomedical prescriptions are very useful to build a consolidated summary for patients.1,2 Pharmaceutical data is an integration of prescription names and other medication information, for example drug name, dosage, course, etc. A significant amount of work has been done in the field of clinical research for clustering of pharmaceutical records for mining useful text. Zhang et al3 worked on ontology-based learning and proposed nine similarity measures of words in clinical document clustering. The ontology based work discussed in4 can help to build an ontology based clinical retrieval system. Patterson et al5 proposed a clustering technique and applied to a number of 17 biomedical notes with the help of an unsupervised clustering algorithm using various lexical and semantic patterns for proper extraction. The work in 6 ontology based access control mechanism is used to secure clinical data.
Han et al 7 clustered clinical notes using latent semantic indexing method and found it as an effective method for measuring the similarity. Nonnegative matrix factorization (NMF) framework significantly outperforms the various classical clustering algorithms such as that of bisecting K-means, and hierarchical clustering. 5 Our contribution to the research paper is summarized as:1 An enhanced framework system for extricating symptom/medication names from clinical documents;2 Application of multi-view NMF to effectively use medication/symptom names to improvise the clustering results;3 Extraction based on age 4) Computing accuracy on the basis of word count and tf-idf factor.
Related Work
Over the past few years, researchers have carried out various works relating to clustering of biomedical notes this section of document presents some of them. Huang et al8 proposed the ensemble non-negative matrix factorization technique for effective clustering of clinical notes. A combination of ensemble non-negative matrix factorization and Hybrid Bipartite Graph formulation (HBGF) is proposed for clustering the prescriptions. Ling et al9 Build a combined approach for extricating drug names and their relating symptom names from a collection of clinical documents. Their work showed a comparison of two approaches namely, non-negative matrix factorization technique and multi-view NMF to produce clusters out of a set of pharmaceutical prescriptions based on different sample-feature matrices generated. Yan et al11 Introduced an efficient multi viewpoint-based similarity measure for clustering clinical notes. This concept was applied on clustering which the authors named as MVSC. Subsequently, they represented it as MVSC-IR and MVSC-Iv i.e. MVSC as a criterion function IR and Iv respectively. The main goal is to perform document clustering by optimizing IR and Iv.
Chim and Deng 12 presented their work using an expression (phrase) based record clustering. They focused their work to compute the similar documents by the usage of a method called as Suffix Tree Documents (STD) model. The authors considered three kinds of suffix namely the root node, leaf nodes, and internal nodes for every document. STD algorithm was then applied to obtain better clustering results than the conventional algorithms. Cheng and Wei 13 A technique named as clustering-based Category-Hierarchy Integration (CHI) was proposed which is an improvement in the technique of Clustering-based Category Integration (CCI) approach. Their execution of category-hierarchy integration showed improvement in the results as compared to the accuracy of that obtained by non-hierarchical category-integration techniques. Hsu et al 14 Stated that knowledge from the clinical sources can be utilized to mine the useful data and also analyze clinical data present in the dataset improving the accessibility to various portions of the record. They extricated the features from the biomedical records, which were mapped to the concepts available in the text data thereby computing results based on concepts mentioned in the knowledge bases. Shehata et al15 presented a model that computed the similarity by calculating the similarity between the sentences present in the documents by analyzing their meaning. Concept-based Analysis was the proposed algorithm that used the concept of likeliness to measure the values of term level, sentence level (ctf), document (tf), and corpus levels (df) for a set of documents. Concept based similarity was applied on various datasets and the results proved that the proposed work outperformed the conventional analysis methods. Zhang et al16 devised a novel approach to enhance the search results for electronic search records by calculating temporal similarity. They proposed an algorithm to combine textual and temporal relevance with the help of adapted hierarchical clustering method for the purpose of re-ranking of healthcare records. This is used for re-ranking and re-positioning of records.
Bagirovet et al17 modified the existing modified k-means algorithm by calculating the cluster in a stepwise increment manner. For this they generated the starting points with the help of the auxiliary functions. The minimization of the function is achieved by applying k-means algorithm on it. Botsis et al18 proposed to apply various clustering algorithm to document networks on the respective values of threshold to obtain document clusters. The authors applied three clustering algorithms namely k-means, visualization of similarities and Louvain to the obtained networks and calculated the performance to determine cluster values. Arthur et al19 applied their technique to achieve a better running time for the k-means approach. The authors discovered that the running time of k-means clustering algorithm was limited by a polynomial of 1/σ and n. The authors concluded that they would like to evaluate the quality of the local optimum obtained using k-means approach and whether the values achieved could be considered as global optimum or not.
Babashzadehet et al20 utilized semantic data to enhance the performance results of clinical IR framework by defining queries in a representative and significant manner. A dataset namely TREC was used for validation of their approach. Results demonstrated the devised approach incomparably improved the performance values of the retrieval of information as compared to conventional keyword-based IR model.
Cao et al21 Presented their technique named as FacetAtlas, which is a multifaceted visualization approach used for visually evaluate rich text datasets. In order to extract the local as well as global values the authors devised an integrated approach by the application of searching technique on advanced visual analytical device. Omiecinskiet and Scheuermann22 presented an efficient parallel approach for record clustering. This technique was run on a SIMD machine. The authors proved that there does not exist any difference between the SplitMerge algorithm and their performance results. Honigman et al 23 work focused on the detection of the ADEs in the patients’ record using a lexicon device. This device was applied on clinical documents to extract Adverse Drug Events (ADEs). Their approach determined various problems in outpatients in an efficient and economic manner. Hripcsak et al24 focused their work on extraction of information regarding patients with tuberculosis using an natural language processing (NLP) method. Also there is an NLP based generic framework discussed in 25 that can help to retrieve the clinical data efficiently. The authors of24 devised a clinical policy to ensure immediate solution to the problem and to isolate these patients. When combined with automated protocols the Clinical protocols produced an extraordinary good results rather than using it alone.
Apostolova et al 26 proposed a technique that automatically segmented medical documents into semantic sections. They used handcrafted rules to develop a scalable biomedical documents segmentation approach that required very less user effort for efficient extraction of information from clinical texts. They used it for automatically identifying high confidence training set. Guan et al 27 used an Affinity Propagation (AP) based approach named as Seeds Affinity Propagation (SAP) to improvise the semi-supervised clustering approach. A dataset, namely the Reuters-21578 from UCI Machine Learning Repository, was selected for comparisons with two other clustering algorithms, namely, AP algorithm and k-means algorithm.
Methodology
Clinical Documents
Clinical documents consist of medical data about patients such as demographic, symptoms, medicines, billing, gender, age or other historical information. The information present is in the unstructured text form. They consist of tags, stem words etc that needs to be removed to a structured format of text. An example of free text format of clinical document is shown in Figure 1.
Pre- Processing
The clinical prescriptions consist of the structured sections which consist of unorganized pharmaceutical information. Enormous amount of hidden medical information can be mined from them by applying right technique. In this research an efficacious approach is employed to pre-process documents in order to extract the valuable words and proper sections from the clinical notes. This results into improvement of the quality of prescriptions present in the dataset. An overview of the pre-processing approach for the extraction of symptoms and medication is described in Figure 2.
 |
Figure 1: An example of textual clinical prescription.
|
Pre-processing of the clinical prescriptions includes the removal of unnecessary words such as stem or stop words, eliminating unnecessary sections from the clinical notes. For this the clinical notes were first pre-processed the text document using the Standard CoreNLP Tool (http://stanfordnlp.github.io/CoreNLP/) which is a java based tool that helps to parse, identify the entities, sentimentally analyse the text.
A number of sections are present in the biomedical documents, symptoms and medication names needs to be extracted from these sections. Majorly the symptom names are contained in sections such as Chief Complain, History of illness, Assessment plan, Physical Exam, Specimen, Review of systems. Likewise, the drug names can be found in the following sections Medication, Impression, Recommendations, Past medical history, Assessment plan, medication on discharge. For such computations section annotator is used in order to differentiate between the sections present in the textual prescription. The header information for the respective sections is computed and based on that the necessary sections are retrieved from the document. The sections which provide the negation recommendations are excluded using the negation annotator. This is done using the NegEx (https://healthinformatics.wikispaces.com/NegEx+Algorithm), a natural language processing (NLP) tool which detects the negative terms present in the clinical text and removes the corresponding medicine name so that right medication is retrieved. This tool identifies the trigger terms and works according to the scope of the terms identified. This tool recognizes the pre-negation and post-negation words such as keptoff, avoid, away, without and was ruled out, free respectively. The clinical note shown in the Figure 1 consists of chief complaint of chest pain. The statements “The naprosyn is not helping that much” and “keptoff Protonix” have negation words “not helping much” and “keptoff” respectively so the negation medicine relating these medicines are removed. Likewise the negation terms such as ‘avoid’, ‘allergic’ are annotated in the document and the corresponding medication is discarded for correct medical recommendation.
After the pre-processing process is over the sample matrices were obtained which has the attributes as shown in Table 1.
Table 1: Size of the Sample Matrices from the Dataset.
| Attributes | #Size |
| Symptom names | 216 |
| Medicine names | 156 |
Symptom/Medication Names Extraction
The medication and symptom names are present in the different sections of the clinical notes. After pre-processing of documents MedEx 28 is used for identifying and extricating the medicine names and MetaMap 10,29 is used for symptom names extraction from these sections. Figure 2 shows the extraction of the same using MedEx and MetaMap after pre-processing the clinical notes. MedEx (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2995636/figure/fig1/)30 is a java based open source tool that helps to identify the medicine terms such as dosage, intake time, drug names, duration and amount in the clinical text. It maps the medication information found to the UMLS thesaurus concepts to find the most precise match for accurate medication extraction. The symptoms extraction is done using MetaMap (https://metamap.nlm.nih.gov/), which is a configurable program that helps to relate the biomedical data to Meta-thesaurus concepts. It uses semantic knowledge representation NLP based approach for mapping the concepts such as “aapp”, “clna”, “clnd”, “nnon” which means amino acid, clinical attribute, clinical drug and nucleic acid respectively. Some other are bact, bodm, enzy, impo, vita etc (https://mmtx.nlm.nih.gov/MMTx/semanticTypes.shtml).
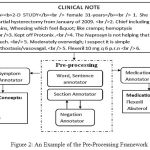 |
Figure 2: An Example of the Pre-Processing Framework
|
Matrix Factorization Technique (M-NMF)
Non-Negative Matrix Factorization
NMF is an efficient method to factorize a given non-negative matrix says, N x M into the product of two non-negative matrices of lower dimension in the forms as matrix N x k and another matrix K x M, which can be efficiently expressed using the Euclidean distance formula 1:
![]()
The NMF aims at finding a low rank estimate of a matrix V (N x M) by considering V as a product of two-dimensional decreased matrices W and H. Each of the columns of W represents a basis vector and that of H contains an encoding of the linear composition of the basis vectors that approximates the respective column of V. Measurements of W and H are N x K and K x M separately, where k is the decreased rank matrix. 9 An efficient technique to update rules of W and H multiplicatively is known as the multiplicative method (MM). The algorithm is stated below as:
Initialization of H and W with non-negative values.
Iterate for each variable c, i, and j until convergence or after l iterations:
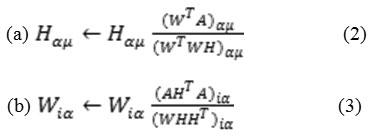
The optimization of Ai ≈ Wi Hi for each view I can be achieved by the equation:
![]()
Multi-View Point NMF
NMF has been effectively utilized in multi-view learning. Multi-view technique helps to identify latent components in various distinct sub-matrices in a simultaneous manner. Liu et al [31] proposed the extension of the basic NMF in an optimized form using different views.
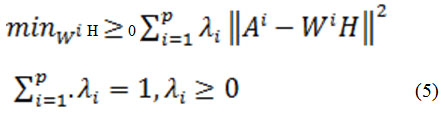
Figure 3 shows the M-NMF technique that utilizes three views i.e. words, symptoms and medication to compute the feature matrix. The matrix is used for the computation of multi-view point similarity computation. As more than one view are utilized the accuracy of the computations made becomes high. Also, the extraction is based on the age of the patients’, so the accuracy value is much higher.
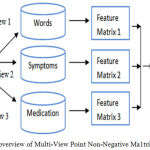 |
Figure 3: An overview of Multi-View Point Non-Negative Ma1trix Factorization
|
Results and Discussion
Dataset Result
The result for the medicines and symptoms extraction is shown in Table 2. The idea is to extricate the medicine and symptoms based on the age of the patients’ using the proficient clustering technique i.e. Multi-View Point Non-negative matrix Factorization (M-NMF). The value of k (cluster value) is taken as three (k=3) for clustering the documents into three clusters.
Table 2: Extraction of symptom and medicine names
| Symptoms | Medicines |
| 1. Chest pain, wheezing, haemoptysis, quot, cramps, moderately overweigh, orthostasis | Flexeril; Albuterol; hcl
|
| 2. Abdominal pain, obstipation, sinus, abdominal scars, Vault prolapsed
|
Nasonex; Xopenex;
Advair |
| 3. Heart failure, fatigue, fever, coronary artery disease, vomiting | Methyldopa; thera;
Dipyridamole |
Evaluation Metrics
Accuracy based on three views is computed in this research work. It is the measurement of the fraction of documents that are labelled correctly. A one-to-one correspondence exists between the true classes and the assigned clusters. Accuracy is calculated using formula 6.
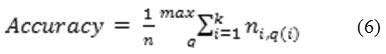
where q is the possible permutation from 1 to k.
Two age groups are considered for the extraction of symptoms and medication names. Table 4 shows the results for the accuracy based on patients’ age for the age group less than 30. As shown in Table 3 the count based on words in the clinical notes is calculated using the feature matrix thereby calculating the accuracy using the formula specified in equation 6. The accuracy based on count and TF-IDF factor as per the respective view is computed.
Table 3: Dataset Results for the Age Group below 30.
| Count | Words | 70.9090909090909 |
| Symptoms/Medicine | 80.7272727272727 | |
| All views | 83.7272727272727 | |
| TF-IDF | Words | 73.4545454545454 |
| Symptoms/Medicine | 78.7272727272727 | |
| All views | 80.9272727272727 |
Table 4: Dataset Results for The Age Group Above 30.
| Type | Views | Accuracy |
| Count | Words | 74.9090909090909 |
| Symptoms/Medicine | 72.7272727272727 | |
| All views | 75.7272727272727 | |
| TF-IDF | Words | 71.4545454545454 |
| Symptoms/Medicine | 76.7272727272727 | |
| All Views | 78.9272727272727 |
Table 4 shows the results for the accuracy based on patients’ age for the age group above 30. As shown in Table 3 the count based on words in the clinical notes is calculated using the feature matrix thereby calculating the accuracy using the formula specified in equation 6. The accuracy based on count and TF-IDF factor as per the respective view is computed.
Conclusion
In this research work a medical framework is implemented for the extraction of symptom and medicine names from the free-text documents. The system uses pre-processing units i.e. negation annotator, section annotator, word annotator. From the extraction of medicines and symptoms three views from the clinical notes. Multi-View NMF is applied for clustering the clinical notes. The accuracy with respect to three views is computed for extraction of the medicines and symptoms based on the age of the patients’ mentioned in the clinical notes. The age based result obtained for both the techniques i.e. NMF and M-NMF showed improved accuracy for M-NMF as compared to NMF technique. It indicates that the M-NMF technique is an improvised version of the NMF and has the capability to perform faster to obtain better results.
Funding Source
No financial support was obtained from anywhere for this project.
Conflicts of Interests
Authors have no conflict of interest.
References
- Roque F. S., Jensen P. B., Schmock H., Dalgaard M.,Andreatta M., Hansen T., Søeby K., Bredkjær S., Juul A., Werge T and others. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput Biol. 2011;7(8):e1002141.
CrossRef - Hripcsak G., Bakken S., Stetson P. D and Patel V. L. Mining complex clinical data for patient safety research: a framework for event discovery. J. Biomed. Inform. 2003;36(1):120–130.
CrossRef - Zhang X.,Jing L., Hu X., Ng M., Xia J and Zhou X. Medical document clustering using ontology-based term similarity measures. 2008.
- Vijayarajan V., Dinakaran M and Lohani M. Ontology based object-attribute-value information extraction from web pages in search engine result retrieval,” Smart Innov. Syst. Technol. 2014;27(1):611–620.
CrossRef - Patterson O and Hurdle J. F. Document clustering of clinical narratives: a systematic study of clinical sublanguages. in AMIA Annu Symp Proc. 2011(2011):1099–1107.
- Mohan K and Aramudhan M. Ontology based access control model for healthcare system in cloud computing. Indian J. Sci. Technol. 2015;8(S9):218–222.
CrossRef - Han C and Choi J. Effect of latent semantic indexing for clustering clinical documents. in Computer and Information Science (ICIS), 2010 IEEE/ACIS 9th International Conference on. 2010;561–566.
CrossRef - Huang X., Zheng X., Yuan W., Wang F and Zhu S. Enhanced clustering of biomedical documents using ensemble non-negative matrix factorization. Inf. Sci. (Ny). 2011;181(11):2293–2302.
CrossRef - Ling Y., Pan X., Li G and Hu X. Clinical documents clustering based on medication/symptom names using multi-view nonnegative matrix factorization. IEEE Trans. Nanobioscience. 2015;14(5):500–504.
CrossRef - Aronson A. R. Metamap Mapping text to the umls metathesaurus. Bethesda, MD NLM, NIH, DHHS. 2006;1–26.
- Yan Y.Chen L and Nguyen D. T. Semi-supervised clustering with multi-viewpoint based similarity measure, in Neural Networks (IJCNN), The 2012 International Joint Conference on. 2012;1–8.
CrossRef - Chim H and Deng X. Efficient phrase-based document similarity for clustering. IEEE Trans. Knowl. Data Eng. 2008;20(9):1217–1229.
CrossRef - Cheng T. H andWei C. P. A clustering-based approach for integrating document-category hierarchies. IEEE Trans. Syst. Man. Cybern. A Syst. Humans. 2008;38(2):410–424.
CrossRef - Hsu W., Taira R. K.,El-Saden S., Kangarloo H and Bui A. A. T. Context-based electronic health record: toward patient specific healthcare. IEEE Trans. Inf. Technol. Biomed. 2012;16(2):228–234.
CrossRef - Shehata S., Karray F and Kamel M. An efficient concept-based mining model for enhancing text clustering. IEEE Trans. Knowl. Data Eng. 2010;22(10):1360–1371.
CrossRef - Zhang J., Huang J. X., Guo J and Xu W. Promoting electronic health record search through a time-aware approach. in Bioinformatics and Biomedicine (BIBM), 2013 IEEE International Conference on. 2013;593–596.
CrossRef - Bagirov A. M., Ugon J and Webb D. Fast modified global k-means algorithm for incremental cluster construction.Pattern Recognit. 2011;44(4):866–876.
CrossRef - Botsis T., J. Scott T., Woo E. J and Ball R. Identifying Similar Cases in Document Networks Using Cross-Reference Structures.IEEE J. Biomed. Heal. informatics. 2015;19(6):1906–1917.
- Arthur D., Manthey B and Röglin H. Smoothed analysis of the k-means method. J. ACM. 2011;58(5):19.
CrossRef - Babashzadeh A., Huang J and Daoud M. Exploiting semantics for improving clinical information retrieval. in Proceedings of the 36th international ACM SIGIR conference on Research and development in information retrieval. 2013;801–804.
CrossRef - Cao N.,Sun J., Lin Y. R., Gotz D., Liu S and Qu H. Facetatlas Multi faceted visualization for rich text corpora. IEEE Trans. Vis. Comput. Graph., 2010;16(6):1172–1181.
CrossRef - Omiecinski E and Scheuermann P. A parallel algorithm for record clustering. ACM Trans. Database Syst. 1990;15(4):599–624.
CrossRef - Honigman B., Light P., Pulling R. M and Bates D. W. A computerized method for identifying incidents associated with adverse drug events in outpatients. Int. J. Med. Inform. 2001;61(1):21–32.
CrossRef - Knirsch C. A., Jain N. L., Pablos-Mendez A., Friedman C and Hripcsak G. Respiratory isolation of tuberculosis patients using clinical guidelines and an automated clinical decision support system. Infect. Control Hosp. Epidemiol. 1998;94–100.
CrossRef - Vijayarajan V., Dinakaran M., Tejaswin P and Lohani M. A generic framework for ontology‑based information retrieval and image retrieval in web data. Human-centric Comput. Inf. Sci. 2016;6:1.
- Apostolova E., Channin D. S., Demner-Fushman D., Furst J., Lytinen S and Raicu D. Automatic segmentation of clinical texts. in Engineering in Medicine and Biology Society, 2009. EMBC 2009. Annual International Conference of the IEEE. 2009;5905–5908.
CrossRef - Guan R., Shi X., Marchese M., Yang C and Liang Y. Text clustering with seeds affinity propagation. IEEE Trans. Knowl. Data Eng. 2011;23(4):627–637.
CrossRef - Xu H., Stenner S. P., Doan S., Johnson K. B., Waitman L. R and Denny J. C. MedEx a medication information extraction system for clinical narratives. J. Am. Med. Informatics Assoc. 2010;17(1):19–24.
CrossRef - Doyle M. The Metamap Process: A New Approach To The Creation Of Object Oriented Image Databases For Medical Education. in Engineering in Medicine and Biology Society. 1991;13:1991. Proceedings of the Annual International Conference of the IEEE. 1991;1046–1047.
CrossRef - Sohn S., Clark C.,Halgrim S. R., Murphy S. P., Chute C. G and Liu H. MedXN an open source medication extraction and normalization tool for clinical text. J. Am. Med. Informatics Assoc. 2014;21(5):858–865.
CrossRef - Liu J.,Wang C., Gao J and Han J. Multi-view clustering via joint nonnegative matrix factorization. in Proceedings of the 2013 SIAM International Conference on Data Mining. 2013;252–260.
CrossRef - Dataset :- (https://idashdata.ucsd.edu/community/45).







