G. L. N. Murthy1 and B. Anuradha2
and B. Anuradha2
1Department of ECE, LBRCE, Mylavaram, Andhra Pradesh, India.
2Department of ECE, S.V. University College of Engineering, Tirupati, A.P, India.
DOI : https://dx.doi.org/10.13005/bpj/1288
Abstract
Unclear boundaries as well as misclassification are the significant problems that need to be addressed in many of the medical imaging related problems. In particular,pathological studies need accurate delineation of objects of interest. Further presence of noise and non-clear boundaries deteriorate the performance of segmentation of brain Magnetic resonance images.Extracting any tissue from brain images fundamentally involveseither registration with atlas or complex deformation models. In the current work, all these problems are addressed by merging the clustering approaches with region growing methods to extract most prominent brain tissue, Hippocampus. Structural analysis of Hippocampus plays a vital role in diagnosis of many cognitive related disorders. An edge enhanced Fuzzy C means (EEFCM) algorithm is proposed aimed at extracting the Hippocampus. The results have shown better results when compared to existing approaches in terms of Dice and Jaccard coefficient.
Keywords
Hippocampus; Skull Removal; Misclassification; Skull Stripping; Edge Enhancement
Download this article as:| Copy the following to cite this article: Murthy G. L. N, Anuradha B. Edge Enhanced Fuzzy C Means Algorithm for Hippocampus Segmentation and Abnormality Identification. Biomed Pharmacol J 2017;10(4). |
| Copy the following to cite this URL: Murthy G. L. N, Anuradha B. Edge Enhanced Fuzzy C Means Algorithm for Hippocampus Segmentation and Abnormality Identification. Biomed Pharmacol J 2017;10(4). Available from: http://biomedpharmajournal.org/?p=17217 |
Introduction
With the symptoms of memory loss, difficulty in performing daily activities and vocabulary becoming hard, Alzheimer’s disease is becoming the one of main cause for death if unnoticed in the earliest stages. It is a progressive neurodegenerative disease that destroys the connections in the brain. Severe atrophy of brain will be the consequence of the death of cells in the outer layer of brain resulting in increased space with in the brain. This damage to the brain will result in problems in memory, intelligence, judgment and behavior.It is known that neurons transmit messages between different parts of brain to muscles and organs of the body. These connections will be lost resulting in sever cognitive disorders.
The damage to the brain starts a decade before memory and cognitive problems evident, which is called as preclinical stage. It is very difficult to observe the complex changes going in the brain during this stage.During this stage, healthy neurons are affected due to abnormal deposits of proteins in the form of Amyloid plaques and Tau tangles. This will lead to sever structural deformation of a tiny tissue in human brain, Hippocampus. Shaped in the form sea horse, this tissue is significantly responsible for memory formation and recollection. Significant amount of research has been carried out and still continuing to develop automated algorithms for assisting in solving the burning problem of the world. Further with the advent of medical imaging techniques, several imaging modalities are existing to aid in clinical diagnosis and treatment planning.
Manually segmenting the large volumes of data derived from any of the imaging technique is much time consuming and prone to error. Further, there is large possibility of intra and inter rater variability. However, while developing automated algorithms, proper care must be taken as the tissue of interest, the Hippocampus is tiny as well as has very unclear boundaries.
In many of the neuro imaging studies as well as clinical diagnosis, segmenting the structures of interest is very much critical. Intensity in homogeneity, presence of noise along with partial volume effects will significantly deteriorates the performance of any algorithm meant for identifying the structural atrophy of brain tissues. Much work has been carried in this regard to identify the level of cognitive impairment.
A hybrid algorithm obtained by merging deformable model with region growing techniques was presented in[1]. The knowledge about the position and shape of hippocampus are involved in the process of Hippocampus delineation. Ghanei has proposed a new method in [2] that detects and follows edges based on external forces. The external energy and limiting movements are continuously mapped to improve the modal stability. An algebraic multi grid based algorithm was proposed in[3], that incorporates a priori knowledge through Bayesian formulation. A manually labeled training set is used to estimate the probabilistic information leading to an accurate detection of tissues of interest.Owen T. Carmichael has compared the performance of various algorithms and concluded that Atlas based segmentation is preferable method due to simplicity and ability to tolerate certain amount of hippocampus localization error.
Registration based methods are used to produce geometric deformations meant for automated segmentation as presented in[5].In [6], Hippocampal segmentation is carried out by minimizing the energy derived from manually labeled images. The significance of atlas selection was presented in [7] and the selection of atlases was assessed both qualitatively and quantitatively. Graph cut algorithm combined with atlas based segmentation was presented in [8]. The graph cut is used to find the global minimum. However, the algorithm suffers from the drawback of lower value of similarity index when compared to conventional methods. A detailed study of automated segmentation algorithms to extract Hippocampus was presented in [9].
In the current work, a novel algorithm is proposed that is intended to solve the problems of unclear boundaries as well computational complexity. A significant reduction in computation time as well as complexity in classification is achieved in the preprocessing step where all the non-brain regions were removed.
Materials and Methods
In the current work, 25 MR images, coronal section were randomly picked from OASIS data base [18].All subjects are right handed with varying age and gender. The selected images cover both normal and abnormal cases.
The main motto of the current work is to reduce the complexities involved in the process of Hippocampus segmentation vide Atlas selection, registration. Instead of opting for a reference template and processing through feature extraction and matching, region growing technique is combined with clustering. Even though the clustering will classify tissues into groups with similar characteristics, unclear boundaries may affect the extraction once a seed point is defined. Thus before proceeding ahead with clustering the edges of the image are further sharpened so that there may not be overlapping of regions in the process. Even the gray level intensities of the image are adjusted to nullify the impact of bias field as well as intensity non uniformity. The preprocessing step will involve removing the non-brain regions through modeling of intensity distribution.Fig.1 summarizes various steps involved in the current work. Brain extraction removes the unwanted regions through modeling of histogram. However as when required based on the level of noise present in the image, a work proposed in [21] can be used.
The underlying concepts of unsharp masking are used to enhance the edges in the next step. Fuzzy c means algorithm is used to group the pixels into different regions after which simply selecting a proper value of seed point will result in the required tissue of interest, the Hippocampus.
 |
Figure 1: Work Flow of the proposed algorithm
|
Skull Stripping
The presence of many non-brain tissues will in general affect the clinical diagnosis and treatment planning while performing MR image analysis. Thus it always becomes mandatory to preprocess human brain to delineate between brain from non-brain regions. This step is significant as any error in at this level may result in misclassification. Much research has been carried out in this regard to yield a better solution towards the problem of skull stripping.In [10], a linear volume registration procedure is used to remove non brain regions. Localization of single white matter voxel followed by creating minimum is the methodology used in[11].Any non-uniformity in the intensities is corrected by using surface deformation process. Aaron Carass hasdeveloped [12] a method based on the combination of segmentation and registration.A comparison between region growing approaches and Morphological approaches was presented in [13]. A deformable model was used [14] to remove non brain regions, where image local gray levels are used control the deformation which is further supported by gray level modeling. A histogram based analysis is used in [15] to solve the problem of presence of non-brain regions. However, while dealing with abnormal images, parameter adjustment is needed to maintain the performance of the algorithm always better. Doshi[16] has addressed the problem of variability between different imaging modalities and proposed a multi atlas based skull removal algorithm.
The current work will utilize the work proposed in [17] to remove the non-brain regions before proceeding for hippocampus segmentation.. The significance of the algorithm is that it is simple and having less computational complexity when compared to other algorithms. The proposed algorithm fundamentally involves three steps: threshold selection through histogram modeling following morphological opening and closing.The main motto behind selecting a threshold is to separate the desired region from unwanted portions. Either it can be back ground including noise or the brain tissues, proper selection of threshold plays key role. The approach involves obtaining the peak difference between probability distribution and normal distribution fit of the given image. This is required as there is need to find the location of separation between both the peaks.The modeled histogram H(x) can be obtained through

Where μ and σ2 are the mean and variance of the image from which skull is to be stripped.
Fig. 2 shows the original image in coronal view taken from OASIS database. Fig.3 indicates the gray level variation of original image with mean gray level value 39.52. After Gaussian modeling, the skull is removed and resulting histogram is shown in Fig.4 and resulting mask is shown in Fig.5.Finally the skull stripped human brain is shown in Fig.6.
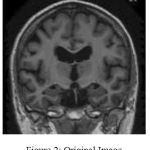 |
Figure 2: Original Image
|
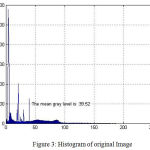 |
Figure 3: Histogram of original Image
|
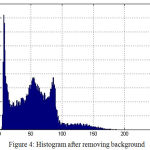 |
Figure 4: Histogram after removing background
|
 |
Figure 5: Generated Mask
|
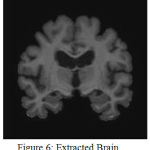 |
Figure 6: Extracted Brain
|
Hippocampus Eextraction and Classification
Human perception is highly sensitive to edges and other fine details in an image and machine perception is not an exception. Smoother or minor variation of gray level data, it becomes cumbersome for any algorithm to carry out the process of delineation of objects of interest .In edge enhancement, at the fundamental level the original image is added to high pass filtered version of the same. This process results in an increase in the contrast between bright and dark regions. The resulting image can be mathematically represented as
![]()
where F(x,y) is the edge enhanced image and is the high pass filtered version of the original image Δ2 f(x,y).
In the current work, once the given MR image is skull stripped, desired edge enhancement is achieved by removing the blurred version of original image from original MR image using the expression,
![]()
where F(x,y) is the edge enhanced image and
![]()
is the low pass filtered version of the original image f(x,y). Fig.7 demonstrates how excellently this step has enhanced the edges of skull stripped image with help of minor gray level adjustments.
Uncertainty is an unavoidably present in medical images, affecting the accuracy of any segmentation algorithm due to the existence of non-clear boundaries and regions. The process of clustering, demarcates the different data points in the given image by grouping them based on pre-defined similarity criterion.A detailed discussion on several clustering methods was presented in [19].Among number of such approaches, fuzzy c means algorithm is a soft clustering technique that allows points in data space to be associated with more than one group.FCM clustering is based on minimizing the objective function defined as
![]()
Where X corresponds to the set of input data points, c number of clusters and U is the member ship matrix. Each element of U is the due weightage given to each point for being allocated to one or the cluster with d being the difference between actual data point and considered cluster center.
The elements of the membership matrix can be derived from the equation given below,

The resulting image is shown in Fig.8.It can be observed from Fig. 9 that enhancing the edges well before clustering has resulted in improved demarcation between boundaries. Once, the points in the data space i.e in the image are clustered the final objective will be extracting the Hippocampus. This is performed by region growing, as it is the most effective and robust method for image segmentation with minimum manual intervention. In the current work, the choice of seed point comprises windowing where the hippocampus region is localized. This is feasible as Hippocampus lies in the lower half section of the coronal MR view. A template is defined with in this localized region that can be considered as seed point. Fig.10 and Fig.11 show the extracted left and right hippocampus respectively. However, improper choice of seed point due to excessive atrophy will result an error resulting in misclassification.
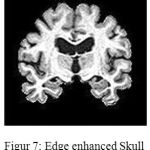 |
Figure 7: Edge enhanced Skull stripped Image
|
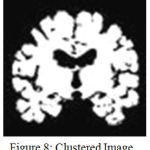 |
Figure 8: Clustered Image
|
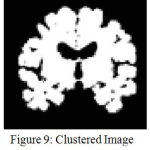 |
Figure 9: Clustered Image without edge enhancemnt
|
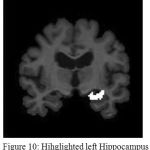 |
Figure 10: Hihglighted left Hippocampus
|
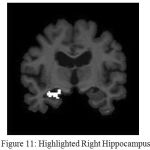 |
Figure 11: Highlighted Right Hippocampus
|
Discussion
Whenever it is desired to develop an algorithm towards clasification of clinical images, much care must must be taken to avoid any error. This is very much needed as any misclassifcation during the process of pre or post segmentaion will have adverse impact on the diagnosis as well as treatment planinng.The developed algorithms should be accurate and computationally less complex. In the current work, concentration is given on both the mentioned parameters, to succesfully segment the Hippcampus and to discrimnate between normal and affected subjects.In this regard as a firststep, processing of insignificant data is avoided by removing all the non brain regions during preprocessing. However, this step can be preceded by noise removal whenver there is a possibilty of image being corruped heavily by noise.A spatial domain depth based non linear algorithm [20] can be used for this purpose.Once noise is removed, the current algoriithm has succesfully addressed the problem of skll stripping that is quite evident from the comparitive resultsalready presented. Table 1 presents a comparison of the proposed work with ref [12] in terms of Dice coefficient. Similarly Table2 is an indicative of how best the proposed algorithm is in terms of Jaccard coefficients. The proposed algorithm performed much better in terms of Jaccard and Dice coefficients.In table 3, the variation in Dice similarity index is compared with a wide range of algorithms. Either average value or minimum and maximum value of Dice coefficient, the proposed algorithm is significantly better than other existing work. These results were represented in Fig.12, Fig.13and Fig.14 respectively.
Intensity in homogentiy and unclear bounaderies are the fundamnetal problem in Hippocampus segmetnation.Once the human brain is skull stripped, before opting for clustering directly edges were enhanced after perfoming the adjustment of intensties.As a consequence of all these steps, stisfactory results were obtained as presented in Fig.15. However, if there exist a severe atrophy of Hippocampus, proper care must be taken unless which there is a possibiity of wrong identification.By considering area as the fundamental parameter, a classification has been made in regard to normal or affected subjects.
Better results were obtained interms of specificity and accuracy when compared to [21].Further the algorithm has shown a better performance when compared to [22] where an effort is made to extract Hippocampus from sagittal section. It was observed that maximum value of Dicecoefficient of 0.899 in the proposed work when compared to 0.87 as obtained in [22]. Even, the results obtained were quite better than the results obtained in [6],[21] [23].
Table 1
| Dice | Mean | Min | Max |
| Ref.12 | 0.9213 | 0.8833 | 0.9445 |
| Proposed Method | 0.9773 | 0.9449 | 0.9793 |
Table 2
| Jaccard | Mean | Min | Max |
| Ref.12 | 0.8547 | 0.7911 | 0.895 |
| Proposed Method | 0.9481 | 0.9449 | 0.9528 |
Table 3
| Dice | Mean | Min | Max |
| BET | 0.931 | 0.894 | 0.967 |
| ROBEX | 0.955 | 0.878 | 0.963 |
| MASS | 0.961 | 0.951 | 0.971 |
| Proposed Method | 0.9773 | 0.9449 | 0.9793 |
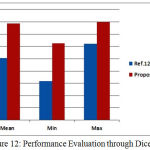 |
Figure 12: Performance Evaluation through Dice
|
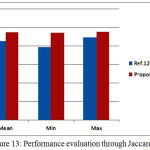 |
Figure 13: Performance evaluation through Jaccard
|
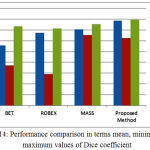 |
Figure 14: Performance comparison in terms mean, minimum and maximum values of Dice coefficient
|
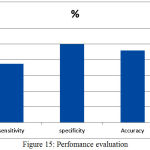 |
Figure 15: Perfomance evaluation
|
References
- Edward A. Ashton, Kevin J. Parker, Fellow, IEEE, Michel J. Berg, and Chang Wen Chen,”A Novel Volumetric Feature Extraction Technique with Applications to MR Images”IEEE Transactions on Medical Imaging, vol. 16, no. 4, august 1997.
- Amir Ghanei , Hamid Soltanian-Zadeh a,b,d, Joe P. Windham,”Segmentation of the hippocampus from brain MRI using deformable contours”,Computerized Medical Imaging and Graphics 22 (1998) 203–216.
CrossRef - Ayelet Akselrod-Ballin, Meirav Galun , John Moshe Gomori, Achi Brandt, and Ronen Basri,”Prior Knowledge Driven Multiscale Segmentation of Brain MRI”,Medical Image Computing and Computer-Assisted Intervention – MICCAI 2007 pp 118-126
- Owen T. Carmichael, Howard A. Aizenstein, Simon W. Davis, James T. Becker, Paul M. Thompson, Carolyn Cidis Meltzer, Yanxi Liu,”Atlas-Based Hippocampus Segmentation In Alzheimer’s Disease and Mild Cognitive Impairment”,Neuroimage. 2005 Oct 1; 27(4): 979–990.
CrossRef - Owen T. Carmichael, Howard A. Aizenstein, Simon W. Davis, James T. Becker, Paul M. Thompson, Carolyn Cidis Meltzer, YanxiLiu,”Atlas-Based Hippocampus Segmentation In Alzheimer’s Disease and Mild Cognitive Impairment”,NeuroImage,2005
- Fadde Van Lijn,Tom Den Heijer,Monique M Breteler,Wiro J Niessen“Hippocampus segmentation in MR images using atlas registration, voxel classification, and graph cuts”,NeuroImage 43 (2008) 708–720
CrossRef - Albert K. Hoang Duc1, Marc Modat, Kelvin K. Leung, M. Jorge Cardoso, Josephine Barnes, Timor Kadir, Sebastein,Ourselin,”Using Manifold Learning for Atlas Selection in MultiAtlas Segmentation”,PLoS One. 2013 Aug 2;8(8)
- Kwak K, Yoon U, Lee DK, Kim GH, Seo SW, Na DL, Shim HJ, Lee JM.,“Fully-automated approach to hippocampus segmentation using a graph-cuts algorithm combined with atlas-based segmentation and morphological opening”,
- Kate E. Macdonald & Kelvin K. Leung & Jonathan W. Bartlett & Melanie Blair & Ian B. Malone & Josephine Barnes,”Automated Template-Based Hippocampal Segmentations from MRI: The Effects of 1.5Tor 3T Field Strength on Accuracy”,Neuroinform (2014) 12:405–412
- Rex DE, Shattuck DW, Woods RP, Narr KL, Luders E, Rehm K, Stoltzner SE, Rottenberg DA and Toga AW, “A meta-algorithm for brain extraction in MRI”, Neuroimage, vol. 23, no. 2, (2004) Oct., pp. 62537
- F. Se´gonne, A. M. Dale, E. Busa, M. Glessner, D. Salat, H. K. Hahn and B. Fischl, “A hybrid approach to the skull stripping problem in MRI” , NeuroImage 22, (2004), pp. 1060–1075.
CrossRef - A. Carass, M. B. Wheeler, J. Cuzzocreo, P.-L.Bazin, S. S. Bassett and J. L. Prince, “A Joint registration and segmentation approach to skull stripping”, 4th IEEE International Symposium on Biomedical Imaging, (2007
- R. Roslan, N. Jamil and R. Mahmud, “Skull Stripping Magnetic Resonance Images Brain Images: Region Growing versus Mathematical Morphology”, International Journal of Computer Information Systems and Industrial Management Applications, vol. 3, (2011), pp. 150-158.
- Galdames FJ, Jaillet F, Perez CA,” An accurate skull stripping method based on simplex meshes and histogram analysis for magnetic resonance images”,Journal of Neuroscience Methods, Elsevier, 2012, 206 (2), pp.103-119
CrossRef - A. G. R. Balan, A. J. M. Traina, M. X. Ribeiro, P. M. A. Marques and C. TrainaJr,, “Smart histogram analysis applied to the skull-stripping problem in T1-weighted MRI”, Computers in Biology and Medicine, vol. 42, (2012), pp. 509–522.
CrossRef - Doshi, G. Erus, Y. Ou, B. Gaonkar and C. Davatzikos, “Multi-Atlas Skull Stripping”, Academic Radiology, vol. 20, no. 12, (2013), pp. 1566-1576.
CrossRef - G. L. N. Murthy, and B. Anuradha,”Non Brain Region Removal in Coronal MR images with Histogram Modeling”,International Journal of Software Engineering and Its Applications Vol. 10, No. 12 (2016), pp. 279-286 http://www.oasis-brains.org/
CrossRef - TamalikaChaira, Medical Image Processing: Advanced Fuzzy Set Theoretic Techniques,CRC Press, 2015.
- Murthy, G. L. N., and Anuradha, B. (2017). An Euclidean Norm Based Nonlinear Filter for Noise Removal In MR Images. i-manager’s Journal on Image Processing, 4(1), 16-22.
CrossRef - K. Somasundaram, T. Genish, T. Kalaiselvi ,”An atlas based approach to segment the hippocampus from MRI of human head scans for the diagnosis of Alzheimer’s disease”, International Journal of Computational Intelligence and Informatics, Vol. 5: No. 1, June 2015.
- G.L.N.Murthy,Dr.B.Anuradha,”Bias field corrected Hippocampus segmentation using k means clustering and region growing”, International Journal of Hybrid Information Technology,Vol.9 No12(2016),pp.47-54
CrossRef - Amoroso N, Errico R, Bruno S, Chincarini A, Garuccio E, Sensi F, Tangaro S, Tateo A, Bellotti R,” Hippocampal unified multi-atlas network (HUMAN): protocol and scale validation of a novel segmentation tool”, Phys Med Biol. 2015 Nov 21;60(22):8851-67.
CrossRef







