Essam Fadel Al-Jumaily and Nagham Qasim Abd
Biotechnology Department Genetic Engineering and Biotechnology Institute for postgraduate studies, University of Baghdad, Iraq.
Corresponding Author E-mail: Samgen992003@Yahoo.com
DOI : https://dx.doi.org/10.13005/bpj/1255
Abstract
Sixty isolates of Pseudomonas aeruginosa were collected from different hospitals of Baghdad. Detection of mexB gene in the isolates was done by using PCR technique and the gene was found in all of the isolates.The susceptibility of the isolates was tested against Carbenicillin, Levofloxacin and Erythromycin by using disc diffusion method.Results showed that 80% of the total isolates was resistant to Carbenicillin while 6% and 32% of the total isolates was resistant to Levofloxacin and Carbenicillin respectively.Three isolates that were resistant to the three antibiotics were chosen to be the base for detecting the gene expression level of mexB before and after the addition of Quinoline-2-one Derivatives.
Keywords
Carbenicillin; Erythromycin; Levofloxacin; mex Bgene; Pseudomonas aeruginosa; Quinoline-2-one Derivatives
Download this article as:| Copy the following to cite this article: Al-Jumaily E. F, Abd N. Q. Effect of Quinoline-2-One Derivatives on the Gene Expression of Mexb of Pseudomonas Aeruginosa. Biomed Pharmacol J 2017;10(3). |
| Copy the following to cite this URL: Al-Jumaily E. F, Abd N. Q. Effect of Quinoline-2-One Derivatives on the Gene Expression of Mexb of Pseudomonas Aeruginosa. Biomed Pharmacol J 2017;10(3). Available from: http://biomedpharmajournal.org/?p=16484 |
Introduction
Pseudomonas aeruginosa is a Gram-negative aerobe that has the ability to infect a wide range of plant and animal host.1 The bacteria is responsible for fatal chronic lung infections in patients with cystic fibrosis and serious acute infections in immunocompromised and injured patients.2
Pseudomonas aeruginosa is one of the curses of the burn units since it is one of the most frequent sources wound and burn sepsis3,4
Antimicrobial agents are expelled from the cells by drug-efflux pumps, which are membrane transporter proteins.5 Efflux pumps expel different types of antibiotics and chemicals such as dyes, detergents, organic solvents and biocides.6
Pseudomonas aeruginosadecreases its susceptibility to antibiotics mainly by the production of RND efflux pump and MexXY-OprM.7 RND family is one type of efflux systems that are commonly found in Gram-negative bacteria.Pseudomonas aeruginosa is of clinical significance due to its ability to acquire high level multi drug resistance and due to its innate multi drug resistance.8 The aim of this study was to evaluate the effect of Quinoline-2-one Derivatives on the gene expression of mexB of Pseudomonas aeruginosa
Materials and Methods
Bacterial Isolates
The isolates were recovered from blood, burn, ears, sputum, urinary tract, catheters and wound from patient admitted to Iraqi medical centers in Baghdad during the period from the sixteenth of October 2016 to the first of February 2017. All of the isolates were identified by their colony characteristics and confirmed by the biochemical test using Api 20 E system
Molecular Detection of Mex B Gene Using PCR Technique
All of the isolates were submitted to PCR technique to detect mexB gene. PCR was performed with a final volume of 20 µl. Each reaction contained 10 µl of GoTaq® Green Master Mix, 1 µl of forward primer, 1 µl of reverse primer, 2 µl of template DNA and 5 µl of nuclease free water. The reaction was submitted to the PCR conditions for 30 cycles and as follow:
Initial denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 seconds, annealing at 60°C for 15 seconds and polymerization at 72°C for 7 minutes. The amplified products were detected by agarose gel electrophoresis. DNA marker (Promega/USA) was run with each gel and genotype was determined by the size of the amplified product.
Antibiotic Susceptibility Testing
Susceptibility of Pseudomonas aeruginosawas tested by disc diffusion test against Carbenicillin, Levofloxacin and Erythromycin, according to clinical and laboratory standards institution (CLSI)guide lines
Antimicrobial activity of Quinoline-2-one Derivatives was determined by agar well diffusion test. The test was carried by inoculating bacterial suspension (1.5 x 108) CFU/mlobtained from (0.5) McFarland standard solution for turbidity. Sterilized loopful was used to inoculate the Muller Hinton agar plates with bacterial suspension by streaking method. Excess moisture was removed by air-dried in sterilized Hood, wells have been made with appropriate depth to have 50 µl of dissolved compounds.
RNA Isolation and Quantitative RT-PCR
RNA was extracted by Trizol reagent from cells grown to the post exponential phase at 37°C. PCR was performed with final volume of 20 µl, each reaction contained 10 µl of GoTaq® Green Master Mix, 0.4 µl of Go Script RT Mix for 1-step RT-qPCR mix, 2 µl of forward primer, 2 µl of reverse primer, 4 µl of RNA and 1.6 of nuclease free water. Reaction was submitted to the PCR conditions for 35 cycles and as follow:
cDNA synthesis at 37°C for 15 minutes, initial denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds and polymerization at 72°C for 30 seconds.
Table 1: Primers used to detect mexB and rpsL genes among Pseudomonas aeruginosa
| Primer | Sequence | Reference | Annealing temp °C | Product size bp |
| Mex B | F 5´-ATCCGCCAGACCATCGCCA-3´
R 5´- CATCACCAGGAACACGAGGAGG-3´ |
Vettoretti et al.,(2009) | 60 | 150 |
| RpsL | F 5´-GCAAGCGCATGGTCGACAAGA-3´
R 5´-CGCTGTGCTCTTGCAGGTTGTGA-3´ |
Dumas et al .,(2006) | 60 | 201 |
Results
A total of 60 clinical isolates of Gram-negative bacteria identifies as Pseudomonas aeruginosa were collected from different sources and as follow: 1 isolate from blood, 18 isolates from burn, 16 isolates from ears, 10 isolates from sputum, 2 isolates from urinary tract, 11 isolates from wound and 2 isolates from catheters (table 2). All isolates were identified through morphological, cultural, biochemical tests and by using Api 20 E system
Table 2: Distribution of Pseudomonas aeruginosa according to the source of samples.
| Source of isolates | No. of isolates | Percentage% |
| Blood | 1 | 1.7% |
| Burns | 18 | 30% |
| Catheters | 2 | 3.3% |
| Ears | 16 | 26.7% |
| Sputum | 10 | 16.7% |
| UTI | 2 | 3.3% |
| Wound | 11 | 18.3% |
| Total | 60 | 100% |
Susceptibility of isolates was tested against Carbenicillin, Levofloxacin and Erythromycin by using disc diffusion test, results showed that 80% of the total isolates were resistant to Carbenicillin, 6% of the total isolates were resistant to Levofloxacin and 32% of the total isolates were resistant to Erythromycin. The majority of isolates of Pseudomonas aeruginosa exhibit intrinsic resistance to many antibiotics because of: the poor outer membrane permeability, constitutive expression of different efflux pumps and production of antibiotic inactivating enzymes.9,10
PCR technique was used to detect mexB gene and the results showed that all of the isolates had the gene. The size of the mexB gene is 150 bp as shown in (Figure1).
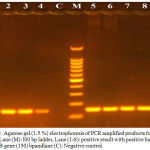 |
Figure 1: Agarose gel (1.5 %) electrophoresis of PCR amplified products for mex B gene. Lane (M): l00 bp ladder, Lane (1-8): positive result with positive bands of mex B gene (150) bpandlane (C): Negative control.
|
In this study three isolates that were resistant to Carbenicillin, Levofloxacin and Erythromycin were chosen to measure the gene expression level of mexB before and after the addition of Quinoline-2-one Derivatives by using q RT-PCR technique after the extraction of RNA from the isolates that were inhibited by the tested antimicrobial agents and from isolates that were treated with Erythromycin due to its inhibitory effect on mexAB-OprM and Levofloxacin as trade antibiotic. The results showed that the gene expression decreased in the case of treating the isolates with Erythromycin, while increase with the expression was noticed with 2.6 than that of control when treating the isolates with Levofloxacin and increasing by 0.2 than that of control was noticed in the case of treating the isolates with Q2 as shown in tables.
Table 3: Ct values and fold of gene expression of mexB gene of Pseudomonas aeruginosa that was treated with Q2 and levofloxacin.
| groups | MIC | Iso. No. | ct of H.K.G | ct of T.G. | Δct | ΔΔct | Folding | Average of folding |
| Un
treated |
P.a.38 | 16.1 | 19.98 | 3.88 | 0 | 1 |
1 |
|
| P.a.49 | 15.9 | 22.03 | 6.13 | 0 | 1 | |||
| P.a.50 | 16.2 | 20.41 | 4.21 | 0 | 1 | |||
|
Q2 |
(256)
µg/ml |
P.a.38 | 16.2 | 19.9 | 3.7 | -0.18 | 1.132883885 |
1.188950775 |
| P.a.49 | 16.1 | 21.8 | 5.7 | -0.43 | 1.347233577 | |||
| P.a.50 | 16.11 | 20.2 | 4.09 | -0.12 | 1.086734863 | |||
|
Lev |
(2)
µg/ml
|
P.a.38 | 15.98 | 18.21 | 2.23 | -1.65 | 3.138336392 |
3.682107727
|
| P.a.49 | 15.8 | 19.8 | 4 | -2.13 | 4.377174805 | |||
| P.a.50 | 16 | 18.06 | 2.06 | -1.82 | 3.530811985 |
Table 4: Ct values and fold of gene expression of mex B gene of Pseudomonas aeruginosa that was treated with erythromycin.
| groups | MIC | Iso. No. | ct of H.K.G | Ct of T.G. | Δct | ΔΔct | Folding | Average of folding |
| Un
treated |
P.a.38 | 16.1 | 19.98 | 3.88 | 0 | 1 |
1 |
|
| P.a.49 | 15.9 | 22.03 | 6.13 | 0 | 1 | |||
| P.a.50 | 16.2 | 20.41 | 4.21 | 0 | 1 | |||
|
Ery |
(64)
µg/ml |
P.a.38 | 16.01 | 23.8 | 7.79 | 3.91 | 0.066523136 |
0.099553601
|
| P.a.49 | 16.02 | 24.98 | 8.96 | 2.83 | 0.140632311 | |||
| P.a.50 | 16 | 23.66 | 7.66 | 3.45 | 0.091505356 |
Discussion
A study made by Al-Marjaniet al., (2013)11 showed that 100% of clinical isolates were resistant to Carbenicillin. Brown and Izundu (2004)12 revealed that 82.4% of isolates were resistant to Nalidixic acid and 76.5% were resistant to Kanamycin, also a study done by Akingbadeet al., (2012)13 showed that 92.7% of isolates were resistant to Amoxicillin and 35.5% were resistant to ciprofloxacin.
Multidrug pumps, particularly those represented by the clinically relevant AcrAB-TolC and Mex pumps of the resistance-nodulation-division (RND) superfamily, not only mediate intrinsic and acquired multidrug resistance (MDR) but also are involved in other functions, including the bacterial stress response and pathogenicity. Additionally, efflux pumps interact synergistically with other resistance mechanisms (e.g., with the outer membrane permeability barrier) to increase resistance levels.14
There are many Researcher found that the efflux-mediated resistance has been found in many bacterial genera. Over expression of an efflux system, responsible for reduction in the accumulation of the antibiotic. The Mex efflux pumps of P. aeruginosaare of particular interest because of their exceptionally broad substrate specificity. While 12 potential efflux systems of this family have been identified in the P. aeruginosagenome .15
Conclusion
We can concluded that development of novel antibiotics that can bypass the effects of efflux pumps is still a challenging task. More studies on involved mechanisms and structure-function association of bacterial efflux systems as well as the interactions between the pumps and other resistance mechanisms are highly recommended.
Acknowledgements
The authors, EssamFadel Al-Jumaily and NaghamQasimAbdgratefully acknowledgeGenetic Engineering and Biotechnology Institute for postgraduate studies ( GEBI), University of Baghdad , Iraq.
Conflict of Interest
The authors declare no conflict of interest.
References
- Fernández L., Gooderham J., Bains M., McPhee J.,Wiegand I and Hancock R. Adaptive Resistance to the last hope antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS . Antimicrob Agents Chemother. 2010;54: 3372-3382.
CrossRef - Bonomo R., Szabo D.Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis. 2006;43(2):S49-56.
CrossRef - Al-Jumaily E. F.,Latif A. S., Al-Bayati R. Effect of a new Quinoline-2-one Derivatives (Compound 3) on Purified DNA gyrase from clinical isolate Pseudomonas aeruginosa IOSR. Journal Of Pharmacy. 2016:12:12-18.
- Saaiq M., Ahmed S., Zaib M. Burn wound infections and antibiotic susceptibility patterns at Pakistan institute of medical science. World J Plast. Surq. 2015;4:9-15.
- Verchère A., Dezi M.,Adrien V., Broutin I., Picard M. In vitro transport activity of the fully assembled mexAB-OprM efflux pump from Pseudomonas aeruginosa. Nat Commun. 2015;22:6.
CrossRef - Askoura M., Mottawea W.,Abujamel T and Taher I. Efflux pump inhibitors (EPIs) as new antimicrobial agents against Pseudomonas aeruginosa . Libyan J Med. 2011;6:10.
CrossRef - Lister P., Wolter D., Hanson N. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Micro Rev. 2009;22:582-610.
CrossRef - Poole K. Pseudomonas aeruginosa Resistance to the Max. Front Microbial. 2011;2:65.
CrossRef - Latif A. Design and Evaluation of Antibacterial Activity (In silico, In vitro and In vivo) of New Quinoline-2-one Derivatives against clinical Pseudomonas aeruginosa. Ph.D. Thesis, Department of Biotechnology, University of Baghdad. 2016.
- Lambert P. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med. 2002;41:22-26.
- AL-Marjani M.,Al-Ammar M and Kadhem E. Occurrence of ESBL and MBL genes in Pseudomonas aeruginosaand Acinetobacter baumannii Isolated from Baghdad, Iraq. Inter. J of Current. Res. 2013;5:2482-2486.
- Brown P., Izundu A. Antibiotic resistance in clinical isolates of Pseudomonas aeruginosain Jamaica. Rev Panam Salud Publica. 2004;16:2
CrossRef - Akingbade O., Balogun S., Oja D., Afolabi R., Motayo B., Okerentugbu P., Okonko I. Plasmid profile analysis of multidrug resistant Pseudomonas aeruginosa isolated from wound infections in South West,Nigeria.World J Appl .sci. 2012;20:766-775.
- Li X. Z., Plésiat P., Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol Rev. 2015;28:337-418.
CrossRef - Mesaros N., Glupczynski Y., Avrain T., Caceres N. E., Tulkens P. M., Van Bambeke F. A combined phenotypic and genotypic method for the detection of Mex efflux pumps in Pseudomonas aeruginosa. Journal of Antimicrobial Chemotherapy. 2007;59:378–386.
CrossRef








