Manuscript accepted on :May 08, 2017
Published online on: --
Plagiarism Check: Yes
Diksha Pandey, Usha Chouhan and Neha Verma
Department of Mathematics, Bioinformatics and Computer Applications Maulana Azad National Institute of Technology, Bhopal 462003, India.
Corresponding Author Email: dikshapandey28@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1164
Abstract
This article provides a comprehensive recap of research on the basis of HIV infection, which is broad in worldwide. Human immunodeficiency virus (HIV) attacks the body's immune system, which is spread through the exchange of body fluids and unprotected sexual contact. HIV is extremely widespread diseases and results from a combination of defects in receptors assays and enzymatic assays, either of which may predominate. There are 33.3 million people worldwide, who suffered with HIV infection. The current prevalence of HIV infection is 0.26% of the India’s population and 0.8% in some countries. The present review gives a brief introduction to various stages, complications, therapeutic targets for receptors and enzymes and obtainable drugs which may activate or inhibit the receptors and enzymes responsible for causing AIDS.
Keywords
Human immunodeficiency virus; Receptors; Enzymes; Drugs; AIDS
Download this article as:| Copy the following to cite this article: Pandey D, Chouhan U, Verma N. HIV Infection: A Review of Their Inhibitors Progression. Biomed Pharmacol J 2017;10(2). |
| Copy the following to cite this URL: Pandey D, Chouhan U, Verma N. HIV Infection: A Review of Their Inhibitors Progression. Biomed Pharmacol J 2017;10(2). Available from: http://biomedpharmajournal.org/?p=15066 |
Introduction
Human immunodeficiency virus (HIV) is initiated from monkey to humans. Monkeys are bringing a virus which is similar to HIV that is called Simian immunodeficiency virus (SIV) (Simonsen, Cameron et al. 1988), (Zhu, Korber et al. 1998). HIV is responsible for Acquired immune deficiency syndrome (AIDS) which is containing a long and endemic history since 1981. AIDS is first acknowledged in the United States (1991) among homosexual and it is spreading more black community compared with white. AIDS basically as HIV grows in an infected individual, it damages specific immune cell, weakening the immune system and allowing the person exposed to infections and illnesses ranging from pneumonia to cancer (Thomas and Quinn 1991). HIV-1 and HIV-2 are the main categories of HIV. A Subspecies of chimpanzees established by native to west equatorial Africa as the original source of HIV-1 which is the virus responsible for the global AIDS pandemic (Gao, Bailes et al. 1999). In Western Africa HIV-2 virus is examined and is seen in other countries as well. It is less infectious and shows progression slowly as compared to HIV-1, although commonly used antiretroviral drugs are dynamic against HIV-2 (Campbell-Yesufu and Gandhi 2011), (Ekouevi, Tchounga et al. 2014). The HIV-1 strains can be separated into four groups such as M, N, O and P. One of this Group such as group M is the first identified and most important group which is responsible for the bulk of the worldwide HIV epidemic. A, B, C, D, E, F, H, J, K and CRFs i.e., circulating recombinant form are the subtypes of M. CRFs are the combination of different subtypes genetic material to form a hybrid virus (Hemelaar 2012). Group O is originated in 1990 which is less widespread than Group M and is largely prescribed to Cameroon, Gabon, and neighboring countries (Mauclère, Loussert-Ajaka et al. 1997), (Peeters, Gueye et al. 1997). Group N is recognized in 1998 which is less epidemic than group O (Vallari, Bodelle et al. 2010) whereas Group P is discovered in 2009 and it is found in a Cameroonian woman living in France (Plantier, Leoz et al. 2009). Despite in depth screening, group P has to date solely been known in one other person, additionally from Cameroon (Vallari, Holzmayer et al. 2011). Although members of all of those groups are able of inflicting CD4+ T-cell depletion and AIDS, they clearly disagree immensely in their distribution inside the human population. The global figure people with HIV are set to go up from the current estimate 33 million and 25 million people have perished. The current prevalence of HIV is between 5% and 30% of adults are infected, which mainly found in eastern, central and southern Africa (Rhodes, Singer et al. 2005). In India, 2.1 million people are facing HIV at the complete year of 2013 and have become the third largest country in the world where people are living with HIV (Reddy, Shah et al. 2005). An infection associated with the HIV virus excessively affects young people at the age 16 to 24 years expressing the highest rates of new HIV infection considered for other age groups and young men such as homosexual and transgender women (Phillips, Wohl et al. 2011). HIV associated with the class of viruses known as retroviruses, which transfer genetic information in the form of RNA. HIV infection destroys the immune system and the central nervous system. The main target of HIV infection is the T helper lymphocyte which is essential in the immune system. T helper cell contains CD4 protein on its surface on which the HIV attaches itself to the cell before gaining entry in to the cell. HIV attachment to the host cell via any of these factors seemingly brings viral envelope glycoprotein such as gp 120 and gp 41 into the nearest of proximity with the viral receptor CD4 and coreceptor (CCR5 or CXCR4) mediated by the V3 loop, increasing the potency of infection (Goodenow and Collman 2006). It has initiated the membrane fusion process as the fusion peptide of gp41 inserted into the target cell membrane, pursued by six-helix bundle formation and complete membrane fusion as shown in Figure1 (Orloff, Orloff et al. 1991).
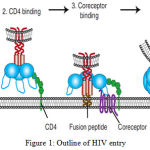 |
Figure 1: Outline of HIV entry |
The process of this virus enters a cell, its RNA is reverse-transcribed to DNA by a first virally encoded enzyme i.e. the reverse transcriptase (RT). This viral DNA enters the cell nucleus in the host genome where it is integrated into the genetic material of the cell by a second virally encoded enzyme called integrase. Activation of the host cell genome that have a consequence transcription of the viral DNA into messenger RNA and it is translated into viral proteins by a third virally encoded enzyme i.e., protease (Ding, Das et al. 1995). This enzyme is prescribed to cleave a viral polyproteins into separate mature proteins. The viral RNA and proteins assemble at the cell surface into new virions and goes into the bud to a cell and released to infect another cell. The extensive cell is damaged from the degradation of the host’s genetic system to the budding and release of virions bulges to the death of the infected cells (Brik and Wong 2003). This article goes over the stages, complications, enzyme involved and receptors of their various inhibitors and treatment.
Stages of HIV Infection
HIV infection has a well-documented development. HIV will eventually overwhelm your immune system. It leads to the being diagnosed with Acquired Immune Deficiency Syndrome (AIDS). However, when the patients are used consistently antiretroviral therapy (ART), impedes the HIV virus from extending and wrecking the immune system. It assists in retaining the body strength by helping fight off life-threatening infections and preventing HIV from progressing to AIDS. In addition, research has shown that pandemic antiretroviral therapy can help impede the spread of HIV to others. Patients may progress through these stages at different rates, depending on a variety of factors are shown in Table 1.
Table 1: HIV infection can widely break down into three distinct stages
| Stages | Cause | Symptoms |
| Primary infection/ Acute infection | At this stage, HIV replicates very quickly and releases new virions into the bloodstream occurs in 2 to 12 weeks and is usually accompanied by a short flu-like illness. This process is known as seroconversion (Rosenberg, Pilcher et al. 2015). | · Rash
· Chills · Headache · Fatigue · Night sweats · Loss of appetite |
| Asymptomatic Infection/ Chronic infection | At this stage, the immune system in someone with HIV slowly weakens, but there have no any symptoms. How long this stage lasts depends on how rapidly the HIV virus copies itself, and how the person’s genes influence the way the body handles the virus (Highleyman 2009).
|
· Aching joints and Muscles
· Fevers · Swollen glands · Sore throat · Upset stomach
|
| Symptomatic Infection | At this stage, virus decreases their immune system so much that they promote infections that other people are capable to fight off. This is known as opportunistic infections and AIDS is caused damage to the immune system (Rowley 2014). | · Diarrhoea
· Tuberculosis · Herpes simplex · Cytomegolavirus |
Complications of HIV Infections
Patients associated with human immunodeficiency virus (HIV) infection often cause multiple complications and comorbidities. The acerbity of abnormality in host defense is the primitive purpose of the risk of promoting distinct pulmonary disorders. Previously in the course of HIV infection, when the immune system is not acutely adjusted, respiratory disorders appear that are approximately identical to those in the general population. Opportunistic infections appear only with severe immunodeficiency. The CD4+ lymphocyte count specifies the maximum predictable alternate marker of immune function, risk of HIV amelioration and opportunistic infection. Amount of HIV activity with serum HIV RNA is adopted commonly to compute the reaction to analysis with antiretroviral agents and to stratify patients by risk of disease amelioration. It is also a responsible predictor than CD4+ counts in certain risk of promoting individual HIV-related diseases (Crothers, Griffith et al. 2005).
Tuberculosis (TB)
Tuberculosis is the most frequent opportunistic infection related to HIV and prominent onset of death surrounded by people with AIDS. HIV positive patients associated tuberculosis are lower cognitive to be sputum-positive with X-rays that serve as less cavitation and higher enmeshment of lower lobes. After completion of treatment they are higher cognitive to fail and die prematurely. Multidrug-resistant Tuberculosis strains are becoming more ubiquitous and the treatment is the frequent 3-4 drug regimen (Walls, Bulifon et al. 2015).
Cytomegalovirus
This common herpes virus is transmitted in body fluids such as blood, semen, urine, breast milk and the healthy immune system and weakens the immune system followed by damaging eyes, digestive tract and other organs. Tumor necrosis factor (TNF α) is mediated stimulation of the host cells, prevailing to intranuclear accumulation and the energizing of the cytomegalovirus DNA replication due to the AIDS patients in cytomegalovirus reactivation (Döcke, Fietze et al. 1994). HIV patients who are not capable to control cytomegalovirus replication have an increased danger of HIV disease development and destruction. Cytomegalovirus infections are related with an increased hazard of dying in the AIDS patients this might be due to organ failure, which is combined to the cytomegalovirus end organ disease in both symptomatic and asymptomatic (Spector, Wong et al. 1998) (Lazaro, Coureau et al. 2006).
Candidiasis
Candidiasis is a frequent HIV-associated infection and caused inflammation and a deep, white coating on the mucous membranes of the mouth, tongue, oesophagus or vagina. Oropharyngeal candidiasis is the first sign of HIV infection and about prevalent fungal opportunistic infection in HIV-infected individuals (Barr 1992).
Cryptococcal Meningitis
Meningitis is an inflammation of the membranes and fluid envelop of the brain and spinal cord (meninges). Cryptococcal meningitis is a prevalent central nervous system infection with HIV and propagated using a fungus found in soil. The study of AIDS associated cryptococcal meningitis established no difference in result between the treatment with amphotericin B and treatment with fluconazole and with the negative cerebrospinal fluid cultures in less than 50 percent of the patients (Saag, Powderly et al. 1992).
Toxoplasmosis
This dormant deadly infection is originated using Toxoplasma gondii and a parasite expanse primarily by cats. Infected cats pass the parasites in their stools and the parasites may then expanse to other animals and humans. Toxoplasma gondii is a necessary intracellular disease that involves a great ratio of the world population, which is a well-known induce of illness encompassed by persons with AIDS (Porter and Sande 1992). Both the immunocompetent individuals and HIV-infected patients, Toxoplasma gondii infection typically is passive and remains asymptomatic. Although, patients associated with HIV are at risk for spreading acute toxoplasmosis due to reactivation of the organism if there is CD4+ T-cell count decreases down 100 cells/µL or if this level decreases down 200 cells/µL in the presence of concurrent opportunistic infection or malignancy (Collazos 2003). If not diagnosed and treated expeditiously then the reactivation of latent Toxoplasma gondii infection in patients with AIDS typically demonstrates as cerebral toxoplasmosis,(Jones, Hanson et al. 1999) which can be life threatening.
Cryptosporidiosis
This infection is caused by an intestinal parasite that commonly found in animals and ingestion of contaminated food or water is likewise suffering from cryptosporidiosis. The parasite develops in the intestines and bile ducts that leads to a severe chronic diarrhea in people with AIDS. A few surveys from developing nations have placed social and behavioral risk factors which are implied in the transmission of cryptosporidiosis in HIV infected patients (Sorvillo, Beall et al. 1998), (Pedersen, Danner et al. 1996), (Glaser, Safrin et al. 1998).
Cancers Common to HIV Infection
Kaposi’s Sarcoma
This cancer is few in people not infected with HIV, but common in HIV-positive people. It normally comes out as pink, reddish or purplish lesions on the skin and oral cavity. Kaposi’s sarcoma can also move the inner organs, letting in the digestive tract and lungs. Women associated with HIV infection through sexual contact with bisexual men are more probable to develop Kaposi’s sarcoma compared with women in other HIV transmission groups. Kaposi’s sarcoma has also been described to occur in HIV-negative homosexual men (Krown 1997). Overwhelming growth has been made toward understanding the pathogenesis of Kaposi’s sarcoma and its connection with HIV infection. HIV infection supported to the pathogenesis using causing profound immunosuppression still loss or deterioration of CD4 cell function (Miles 1996).
Lymphomas
This cancer exists in the white blood cells, especially first appears in the lymph nodes. The most prevalent early symptom is painless swelling of the lymph nodes in the neck, armpit or groin. The common HIV-associated lymphomas includes Burkitt’s lymphoma (BL), other less combative non-Hodgkin’s Lymphomas (NHL), diffuse large B-cell lymphoma (DLBCL), primitive effusion lymphoma (PEL), plasmablastic (lymphoblastic) lymphoma of the oral cavity and primitive central nervous system lymphoma (Jaffe 2001), (Mbulaiteye, Biggar et al. 2003).
Other Complications
Wasting Syndrome
Combative treatment regimens have reduced the number of instances of wasting syndrome, but it still incorporates many people with AIDS and leads to a loss of atleast 10 percent of body weight, usually surrounded by diarrhea, chronic weakness, fever etc.
Neurological Complications
However, AIDS does not infect the nerve cells and causes neurological symptoms such as confusion, depression, anxiety and difficulty in walking. The most common neurological complications are AIDS dementia complex, which contributes to behavioral changes and decreased mental performance.
Kidney Disease
HIV-associated nephropathy (HIVAN) is an excitation of the tiny filters in the kidneys that take out spare fluid and wastes from the bloodstream and bring them in the urine. The risk of developing HIVAN is much higher in blacks because of a genetic predisposition.
An Enzyme Involved in HIV Infection
Reverse Transcriptase
Reverse transcriptase (RT) is an enzyme important for the reproduction of HIV causing AIDS. It converts the single-stranded viral RNA to double-stranded DNA which is integrated into the host cell membrane. The enzyme contains two active sites: polymerase site (Pol) and RNase H site which are desired for the virus activity. When a mature HIV-1 virion is infected a vulnerable target cell and their interactions of the envelope glycoprotein with the coreceptors on the surface of the cell transferred about a fusion of the membranes of the host cell membrane and the virion (Wilen, Tilton et al. 2012). The reverse transcriptase enzyme is an asymmetric heterodimeric and it subsists of two polypeptides 66-kDa subunit (p66) and 51-kDa subunit (p51), 66-kDa subunit (p66) having 560-amino-acids and 51-kDa subunit (p51) having 440-amino-acids (di Marzo Veronese, Copeland et al. 1986), (Lightfoote, Coligan et al. 1986). The p51 subunit shows a more structural role whereas p66 subunit is an open conformation such that the thumb rotates away from the fingers to form a large cleft, which contains double-stranded nucleic acid. If the absence of nucleic acid, then the p66 subunit presume a closed conformation, in which the thumb rotates toward the fingers to fill much of this cleft (Hsiou, Ding et al. 1996). β-thujaplicinol is shown space-filled in magenta and red and is the first protein-ligand crystal structure to provide us a glance of an active site RNase H inhibitor bound to full-length HIV-1 reverse transcriptase are shown in Figure 2 (a) (Himmel, Maegley et al. 2009). The HIV-1 infection includes two classes of reverse transcriptase inhibitors which target the viral enzyme with two different mechanisms: nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs/NtRTIs) and nonnucleoside reverse transcriptase inhibitors (NNRTIs). Naturally, the NtRTIs are involved in deoxynucleosides such as adenosine, thymidine, guanosine and cytidine for incorporation into the elongating double-stranded DNA chain whereas NRTIs lack of 3′-OH group on the deoxyribose sugar and it is also responsible to block the action of reverse transcriptase enzyme therefore, it is able to prevent the further synthesis of viral DNA. NNRTIs don’t require intracellular metabolism (Merluzzi, Hargrave et al. 1990) and bind directly to a hydrophobic pocket which is present in near of the catalytic site of HIV-1 RT known as the NNRTI-binding pocket (Das, Clark et al. 2004), resulting in inhibition of HIV replication. In 2007, description of reverse transcriptase enzyme inhibitors has been studied using molecular docking, chemical similarity and MM-GB/SA Scoring in USA (Barreiro, Guimarães et al. 2007).
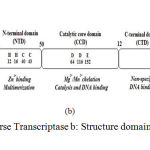 |
Figure 2a: Overview of Reverse Transcriptase b: Structure domains of HIV-1 integrase c: Structure of HIV-1Protease
|
Integrase
Integration of viral DNA into the host cellular DNA is crucial step in the replication cycle of HIV and other retroviruses (Berg and Howe 1989), (Coffin, Tsichlis et al. 1983). HIV integrase can appear approximately at any location in the genome, but certain regions of chromatin are selected (Ciuffi 2008). HIV integrase enzyme is stably maintained and replicated on the cellular DNA over cycles of cell division. HIV integrase has 32KDa proteins with 288 amino acids which encoded by the 39-end region of the pol gene that is cleaved from a polyproteins precursor by the viral protease. HIV-1 integrase composed of three distinct domains such as the N-terminal domain (NTD), the catalytic core domain (CCD) and the C-terminal domain (CTD) are shown in Figure 2 (b). The N-terminal domain (1-49 residues) encompasses conserved pairs of histidine (H12, H16) and cysteine (C40 and C43) residues that bind with zinc and contributes to protein multimerisation of integrase (Zheng, Jenkins et al. 1996), (Lee, Xiao et al. 1997). The catalytic core domain (residues 50-212) is a superfamily of polynucleotidyl transferases having the D, D-35, E motif which are fundamental and conserved between viral integrase and transposases in the catalytic activity (Esposito and Craigie 1998). Metallic cationic cofactor is reconciliated using two residues of the catalytic triad (D64 and D116 for HIV-1 integrase) (Goldgur, Dyda et al. 1998), (Maignan, Guilloteau et al. 1998). The C-terminal domain (CTD) (amino acids 213–288) is rich in basic amino acids and adapts an SH3-like fold (Eijkelenboom, Lutzke et al. 1995) which bind nonspecifically to DNA and therefore it is predominantly concerned with the stability of the complex with DNA. These three domains have been purified, crystallised and characterized in either individually, complex with other proteins or as double-domain partial structure (Li, Krishnan et al. 2011). Growth of target inhibitors require enzyme-specific in HIV integrase. Cell free process was initially developed on the basis of three steps base deletion/transesterification mechanism of the integrase enzyme. In the first step, two nucleotides are excised from each 3′-end of the viral DNA that procedure known as 3′-end processing and in another step, the integration of the processed viral DNA ends into host DNA concerted using integrase catalytic activity, this process is well-known as DNA strand transfer. The HIV-1 integrase catalytic reaction ensues in first two steps of integration. Then, in the third cellular enzymes restoration the single gaps in the DNA chain using eradicating the two unpaired nucleotides at the 5′-ends of the viral DNA. These three reactions also occur in vivo in a consecutive manner and these reactions are also vigorous individually. Compounds have been identified that inhibit integration process which target the active site by an entirely different mechanism (De Luca, Ferro et al. 2010). In 2016, description of HIV-integrase inhibitors identification has been studied structurally using pharmacophore-based virtual screening and molecular dynamics studies in South Africa (Islama and Pillay).
Protease
HIV protease (HIV PR) is unique and viral homodimeric aspartyl protease which cleaves the Gag and Gag-Pol viral polyproteins to mature structural proteins and viral enzymes (DEBOUCK 1992). Polyproteins processing is necessary for the maturation of fully infectious virions. HIV protease has been found as budded immature viral particles that involve catalytically inactive protease cannot undergo maturation to transferable form (Kohl, Emini et al. 1988). There are two highly mobile regions in the HIV protease molecule which are particularly important, the pair of flaps (a glycine-rich loop) are shown in Figure 2 (c) (James, Sielecki et al. 1982) which are seen to close over the substrate-derived inhibitor in the crystal state and one of the two essential aspartyl residues, Asp-25 and Asp-25՛ which lie on the bottom of the cavity. (Martilla and James 1977), (Tourova and Antonov 1988), (Scripture, Voelker et al. 1987). Catalytic water hydrogen-bonded between the two aspartic acids is determined in unliganded structures of monomeric and retroviral aspartyl proteases (Sielecki, Fedorov et al. 1990), (Suguna, Padlan et al. 1987), (Wlodawer, Miller et al. 1989). In order to accomplish their catalysis, these enzymes use two different mechanisms by dividing them into two broad classes of protease enzymes. The first class of enzymes attacks the amide bond carbonyl of the substrate’s scissile bond using an activated water molecule which can be achieved either by a zinc cation (the zinc metalloproteinases) or by the two aspartyl β-carboxy groups at the active site (the aspartate proteases). The second class of enzymes use a nucleophilic atom of an amino acid side chain to initiate amide hydrolysis. The nucleophilic water attacks the carbonyl carbon of the substrate to form a gemdiol inter-mediate. According to several studies, HIV protease inhibitors are able to selectively inhibiting the transport function and this effect may be responsible for a major iatrogenic complication frequently which observed in HIV patients. In 2008, the function and structural of HIV protease inhibitors has been studied using simplicial neighborhood analysis of protein packing (SNAPP) method in USA (Zhang, Kaplan et al. 2008).
Antiretrovial Treatment
The primarily unpredictable number of drugs have been accepted for the therapeutics of the HIV-infected patients (Mehellou and De Clercq 2010). In the treatment of viral infections, armamentarium in different combination therapeutic regimens has transferred a highly lethal syndrome into a chronic disease in effective drugs identification successfully. However, the intendance of the disease is complex and anxious due to the issues such as monitoring of therapy efficacy, chronic administration drug toxicity, drug resistance development, or therapy adjustment after treatment failures. Various steps have been validated as drug targets and finally the number of viral inhibitors has been identified and developed against HIV life cycle (Tsibris and Hirsch 2010). HIV-RNA is an approved alternate for predicting adequacy of antiretroviral and there is no longer needed for the development of antiretroviral therapy. Table 2 summarizes recommended treatment durations to support approvals of indications for the listed groups. The general groups of patient types included in Table 2 take into account both viral susceptibility and treatment history. Patients in Group B and Group C have characteristics that may overlap, decisions regarding the best group to describe a particular individual will be a matter of clinical judgment to some extent. Community drug resistance profiles and the availability of new drugs change over time sponsors may develop protocol inclusion criteria for Group B patients that best describe a multidrug resistant population for which their particular investigational drug would be suitable at the time of enrollment in a protocol.
Table 2: Guidance for adequacy and immunity determination time period, according to HIV-1 Infection Patients
| Patients Groups | Adequacy result of time period | Immunity result of time period |
| Group A: Fully accepted to all approved drugs which gives a treatment spontaneous or previous treatment with a well-documented history determining no virology failure. | Virology response at 48 weeks | Immunity response through 48 weeks |
| Group B: Utilization of multiple drugs and multiple drug categories, but not capable to create a treatment regimen that can suppression of HIV-RNA to levels below assay quantification limits. | Virology response at 24 weeks | Immunity response through 24 weeks |
| Group C: Drug resistance is also present but to able to create a treatment regimen that can suppression of HIV-RNA to levels below assay quantification limits. | Virology response at 24-48 weeks | Immunity response through 24 weeks |
Development of specific entry inhibitors in HIV concentrated on the testing and design of recombinant soluble CD4 molecules. Small molecule inhibitors, which can block the gp120-CD4 interaction that show to lead the promise (Guo, Ho et al. 2003), (Lin, Blair et al. 2003). Ibalizumab binds to the C2 domain of CD4 cell but doesn’t interfere with immunological functions that are involved in antigen presentation (Boon, Holland et al. 2002), (REIMANN, LIN et al. 1997). Aplaviroc is a CCR5 antagonist that determined antiviral activity during short-term monotherapy studies with minimal toxicities and it blocks MIP-1α at nanomolar concentrations, RANTES-mediated signaling is less persuadable to aplaviroc inhibition (Watson, Jenkinson et al. 2005). Various individual drugs have been esteemed by the United States Food and Drug Administration for the treatment of HIV-1 infection. These drugs are distributed into different classes i.e., non-nucleoside reverse transcriptase inhibitors (NNRTI), integrase inhibitors, protease inhibitors (PI) and nucleoside reverse transcriptase inhibitors (NRTI) are shown in Table 3.
Table 3: FDA approved drugs for the treatment of HIV-1 infection
| Drug Name | Generic Name | Class | Approval Date |
| Epivir | Lamivudine, 3TC | NRTI | 17/11/1995 |
| Videx | Didanosine, ddI | NRTI | 09/10/1991 |
| Ziagen | Abacavir, ABC | NRTI | 17/12/1998 |
| Viramune | Nevirapine, NVP | NNRTI | 21/06/1996 |
| Sustiva | Efavirenz, EVF | NNRTI | 17/09/1998 |
| Intelence | Etravirine, ETV | NNRTI | 18/01/2008 |
| Crixivan | Indinavir, IDV | PI | 13/03/1996 |
| Aptivus | Tipranavir, TPV | PI | 02/06/2005 |
| Prezista | Darunavir | PI | 23/06/2006 |
| Isentress, | Raltegravir RTV | Integrase | 12/10/2007 |
| Vitekta | Elvitegravir | Integrase | 24/09/2014 |
| Genvoya | Elvitegravir | Integrase | 05/11/2015 |
Conclusion
The enzymes and receptors which have been studied in this paper are found to be very important for the identification and development of new drugs and therapeutics which can prevent the enzymes from catalytic activity and defend receptors from HIV entry. Many drugs have been found to be successfully inhibit the enzymes but over a certain period these drugs acquired resistance. Further research in this direction may well lead to the development of new allosteric inhibitors of enzymes and receptors that can complement retroviral compounds in treating AIDS patients. The discovery and successful development of HIV overview inhibitors proves viable target for therapeutic intervention in patients. Inhibitors for HIV infection that are believed to have no counterpart in patients are expected to be selective and safe. The competence of HIV to expand drug resistance however, will ultimately outcome in treatment failures to this new class of inhibitors. Though, it is important to understand how resistance expands and what mechanisms the virus is capable for suppression of inhibition. The current research highlighted a starting point towards the HIV virus entry and understanding their complex mechanisms and various inhibitors which has been approved by FDA. The paper also discuss about various complications and risks associated with HIV infection such as Tuberculosis (TB), Cytomegalovirus, Candidiasis, Cryptococcal meningitis, Toxoplasmosis, Cryptosporidiosis, various types of cancers and other disorders. The study of receptor structures, enzyme structures and active molecules for contribution in the anti-HIV drug development has also been briefly described.
Acknowledgement
The authors are highly grateful to the Department of Biotechnology, New Delhi for providing financial support for this work under the Bioinformatics infrastructure facility of DBT at Department of Mathematics, Bioinformatics and Computer Application MANIT Bhopal.
Reference
- Barr C. E. Oral diseases in HIV-1 infection. Dysphagia. 1992;7(3):126-137.
CrossRef - Barreiro G., et al. “Search for non-nucleoside inhibitors of HIV-1 reverse transcriptase using chemical similarity, molecular docking, and MM-GB/SA scoring.” Journal of chemical information and modeling. 2007;47(6):2416.
CrossRef - Berg D. E. and Howe M. M. Mobile DNA, American Society for Microbiology Washington, DC. 1989.
- Boon L., et al. “Development of anti-CD4 MAb hu5A8 for treatment of HIV-1 infection: preclinical assessment in non-human primates.” Toxicology. 2002;172(3):191-203.
CrossRef - Brik A and Wong C. H. “HIV-1 protease: mechanism and drug discovery.”Organic & biomolecular chemistry. 2003;1(1):5-14.
CrossRef - Campbell-Yesufu O. T and Gandhi R. T. “Update on human immunodeficiency virus (HIV)-2 infection.” Clinical infectious diseases. 2011;52(6):780-787.
CrossRef - Ciuffi A. “Mechanisms governing lentivirus integration site selection.” Current gene therapy. 2008;8(6):419-429.
CrossRef - Coffin J. M., et al. “Genomes of endogenous and exogenous avian retroviruses.” Virology. 1983;126(1):51-72.
CrossRef - Collazos J. “Opportunistic infections of the CNS in patients with AIDS.” CNS drugs. 2003;17(12): 869-887.
CrossRef - Crothers K., et al. “The impact of cigarette smoking on mortality, quality of life and comorbid illness among HIV‐positive veterans.” Journal of general internal medicine. 2005;20(12):1142-1145.
CrossRef - Das K., et al. “Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants.” Journal of medicinal chemistry. 2004;47(10):2550-2560.
CrossRef - De Luca L., et al. “Small molecules targeting the interaction between HIV-1 integrase and LEDGF/p75 cofactor.” Bioorganic & medicinal chemistry. 2010;18(21):7515-7521.
CrossRef - DEBOUCK C. “The HIV-1 protease as a therapeutic target for AIDS.” AIDS research and human retroviruses. 1992;8(2):153-164.
CrossRef - di Veronese F. M., et al. “Characterization of highly immunogenic p66-p51 as the reverse transcriptase of HTLV-III-LAV.” Science. 1986;231:1289-1292.
CrossRef - Ding, J., et al. “Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors.” Nature Structural & Molecular Biology. 1995;2(5):407-415.
CrossRef - Döcke W., et al.”Cytomegalovirus reactivation and tumour necrosis factor.”The Lancet. 1994;343(8892):268-269.
CrossRef - Eijkelenboom A. P., et al. “The DNA-binding domain of HIV-1 integrase has an SH3-like fold.” Nature Structural & Molecular Biology. 1995;2(9):807-810.
CrossRef - Ekouevi D. K., et al.”Antiretroviral therapy response among HIV-2 infected patients a systematic review.” BMC infectious diseases. 2014;14(1):461.
CrossRef - Esposito D and Craigie R. “Sequence specificity of viral end DNA binding by HIV‐1 integrase reveals critical regions for protein–DNA interaction.” The EMBO journal. 1998;17(19):5832-5843.
CrossRef - Gao F., et al. “Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes.” Nature. 1999;397(6718):436-441.
CrossRef - Glaser C. A., et al. “Association between Cryptosporidium infection and animal exposure in HIV-infected individuals.” JAIDS Journal of Acquired Immune Deficiency Syndromes. 1998;17(1):79-82.
CrossRef - Goldgur Y., et al. “Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.” Proceedings of the National Academy of Sciences. 1998;95(16):9150-9154.
CrossRef - Goodenow M. M and Collman R. G.”HIV-1 coreceptor preference is distinct from target cell tropism: a dual-parameter nomenclature to define viral phenotypes.” Journal of leukocyte biology. 2006;80(5):965-972.
CrossRef - Guo Q., et al. “Biochemical and genetic characterizations of a novel human immunodeficiency virus type 1 inhibitor that blocks gp120-CD4 interactions.” Journal of virology. 2003;77(19): 10528-10536.
CrossRef - Hemelaar J. “The origin and diversity of the HIV-1 pandemic.” Trends in molecular medicine. 2012;18(3):182-192.
CrossRef - Highleyman L.”Inflammation, immune activation, and HIV.” BETA: bulletin of experimental treatments for AIDS: a publication of the San Francisco AIDS Foundation. 2009;22(2):12-26.
- Himmel D. M., et al. “Structure of HIV-1 reverse transcriptase with the inhibitor β-thujaplicinol bound at the RNase H active site.” Structure. 2009;17(12):1625-1635.
CrossRef - Hsiou Y., et al. “Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms.” Structure 1996;4(7):853-860.
CrossRef - Islama M. A and Pillay T. S. “Structural requirements for potential HIV-integrase inhibitors identified using pharmacophore-based virtual screening and molecular dynamics studies1.”
- Jaffe E. S. Pathology and genetics of tumours of haematopoietic and lymphoid tissues, Iarc. 2001.
- James M., et al. “Conformational flexibility in the active sites of aspartyl proteinases revealed by a pepstatin fragment binding to penicillopepsin.” Proceedings of the National Academy of Sciences. 1982;79(20):6137-6141.
CrossRef - Jones J. L., et al. “Surveillance for AIDS-defining opportunistic illnesses, 1992–1997.” Archives of Dermatology. 1999;135(8):897-902.
CrossRef - Kohl N. E., et al. “Active human immunodeficiency virus protease is required for viral infectivity.” Proceedings of the National Academy of Sciences. 1988;85(13):4686-4690.
CrossRef - Krown S. E. “Acquired Immunodeficiency Syndrome–Associated Kaposi’s Sarcoma: Biology and Management.” Medical Clinics of North America. 1997;81(2):471-494.
CrossRef - Lazaro E., et al. “Change in T-lymphocyte count after initiation of highly active antiretroviral therapy in HIV-infected patients with history of Mycobacterium avium complex infection.” Antiviral therapy. 2006;11(3):343.
- Lee S. P., et al. “Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro.” Biochemistry. 1997;36(1):173-180.
CrossRef - Li X., et al. “Structural biology of retroviral DNA integration.” Virology. 2011;411(2):194-205.
CrossRef - Lightfoote M. M., et al. “Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus.” Journal of virology. 1986;60(2):771-775.
- Lin P. F., et al. “A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding.” Proceedings of the National Academy of Sciences. 2003;100(19):11013-11018.
CrossRef - Maignan S., et al. “Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.” Journal of molecular biology. 1998;282(2):359-368.
CrossRef - Martilla J. A and James J. C. “Importance-performance analysis.” The journal of marketing. 1977;77-79.
CrossRef - Mauclère P., et al. “Serological and virological characterization of HIV‐1 group O infection in Cameroon.” Aids. 1997;11(4):445-453.
CrossRef - Mbulaiteye S. M., et al. “Immune deficiency and risk for malignancy among persons with AIDS.” JAIDS Journal of Acquired Immune Deficiency Syndromes. 2003;32(5):527-533.
CrossRef - Mehellou Y and De Clercq E. “Twenty-six years of anti-HIV drug discovery: where do we stand and where do we go?” J. Med. Chem. 2010;53(2):521-538.
CrossRef - Merluzzi V. J., et al. “Inhibition of HIV-1 replication by a nonnucleoside reverse transcriptase inhibitor.” Science. 1990;250(4986):1411.
CrossRef - Miles S. A. “Pathogenesis of AIDS-related Kaposi’s sarcoma: evidence of a viral etiology.” Hematology/oncology clinics of North America. 1996;10(5):1011-1021.
CrossRef - Orloff G. M., et al. “Penetration of CD4 T cells by HIV-1. The CD4 receptor does not internalize with HIV, and CD4-related signal transduction events are not required for entry.” The Journal of immunology. 1991;146(8):2578-2587.
- Pedersen C., et al. “Epidemiology of cryptosporidiosis among European AIDS patients.” Genitourinary medicine. 1996;72(2):128-131.
CrossRef - Peeters M., et al. “Geographical distribution of HIV‐1 group O viruses in Africa.” Aids. 1997;11(4):493-498.
CrossRef - Phillips G., et al. “Epidemiologic data on young men of color who have sex with men.” AIDS patient care and STDs. 2011;25(S1):S3-S8.
CrossRef - Plantier J. C., et al. “A new human immunodeficiency virus derived from gorillas.” Nature medicine. 2009;15(8):871.
CrossRef - Porter S. B and Sande M. A. “Toxoplasmosis of the central nervous system in the acquired immunodeficiency syndrome.” New England Journal of Medicine. 1992;327(23):1643-1648.
CrossRef - Reddy K. S., et al. “Responding to the threat of chronic diseases in India.” The Lancet. 2005;366(9498):1744-1749.
CrossRef - REIMANN K. A., et al. “A humanized form of a CD4-specific monoclonal antibody exhibits decreased antigenicity and prolonged plasma half-life in rhesus monkeys while retaining its unique biological and antiviral properties.” AIDS research and human retroviruses. 1997;13(11):933-943.
CrossRef - Rhodes T., et al. “The social structural production of HIV risk among injecting drug users.” Social science & medicine. 2005;61(5):1026-1044.
CrossRef - Rosenberg, N. E., et al. “How can we better identify early HIV infections?” Current Opinion in HIV and AIDS. 2015;10(1):61.
CrossRef - Rowley C. F. “Developments in CD4 and viral load monitoring in resource-limited settings.” Clinical infectious diseases. 2014;58(3):407-412.
CrossRef - Saag M. S., et al.”Comparison of amphotericin B with fluconazole in the treatment of acute AIDS-associated cryptococcal meningitis.” New England Journal of Medicine. 1992;326(2):83-89.
CrossRef - Scripture J. B., et al.”High-affinity L-arabinose transport operon: nucleotide sequence and analysis of gene products.” Journal of molecular biology. 1987;197(1):37-46.
CrossRef - Sielecki A. R., et al. “Molecular and crystal structures of monoclinic porcine pepsin refined at 1.8 A resolution.”Journal of molecular biology. 1990;214(1):143-170.
CrossRef - Simonsen J. N., et al. “Human immunodeficiency virus infection among men with sexually transmitted diseases.” New England Journal of Medicine. 1988;319(5):274-278.
CrossRef - Sorvillo F., et al.”Seasonality and factors associated with cryptosporidiosis among individuals with HIV infection.” Epidemiology and infection. 1998;121(01):197-204.
CrossRef - Spector S. A., et al. “Plasma cytomegalovirus (CMV) DNA load predicts CMV disease and survival in AIDS patients.” Journal of Clinical Investigation. 1998;101(2):497.
CrossRef - Suguna K., et al.”Binding of a reduced peptide inhibitor to the aspartic proteinase from Rhizopus chinensis: implications for a mechanism of action.” Proceedings of the National Academy of Sciences. 1987;84(20):7009-7013.
CrossRef - Thomas S. B and Quinn S. C. “The Tuskegee Syphilis Study, 1932 to 1972: implications for HIV education and AIDS risk education programs in the black community.” American journal of public health. 1991;81(11):1498-1505.
CrossRef - Tourova T andAntonov A. “8 Identification of Microorganisms by Rapid DNA-DNA Hybridization.”Methods in microbiology. 1988;19:333-355.
CrossRef - Tsibris A. M and Hirsch M. S. “Antiretroviral therapy in the clinic.” Journal of virology. 2010;84(11):5458-5464.
CrossRef - Vallari A., et al. “Four new HIV-1 group N isolates from Cameroon: Prevalence continues to be low.” AIDS research and human retroviruses. 2010;26(1):109-115.
CrossRef - Vallari A., et al. “Confirmation of putative HIV-1 group P in Cameroon.”Journal of virology. 2011;85(3):1403-1407.
CrossRef - Walls G., et al.”Drug-resistant tuberculosis in HIV-infected patients in a national referral hospital, Phnom Penh, Cambodia.” Global health action 8. 2015.
CrossRef - Watson C., et al. “The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor.” Molecular pharmacology. 2005;67(4):1268-1282.
CrossRef - Wilen C. B., et al. “HIV: cell binding and entry.” Cold Spring Harbor perspectives in medicine. 2012;2(8):006866.
CrossRef - Wlodawer A., et al. “Crystal Structure of Synthetic HIV-Protease.” Science. 1989;245:6I6.
- Zhang S., et al. “HIV‐1 protease function and structure studies with the simplicial neighborhood analysis of protein packing method.” Proteins: Structure, Function and Bioinformatics. 2008;73(3):742-753.
CrossRef - Zheng R., et al. “Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity.” Proceedings of the National Academy of Sciences. 1996;93(24): 13659-13664.
CrossRef - Zhu T., et al. “An African HIV-1 sequence from 1959 and implications for the origin of the epidemic.” Nature. 1998;391(6667):594-597.
CrossRef







