Besharat Rahimi1 and Ahmad Vesal2
1Advanced Thoracic Research Centre, Tehran University of Medical Science, Tehran, Iran.
2Department of Pediatric Cardiology , Rajaie Cardiovascular Medical and Research Center, Iran University of Medical sciences, Tehran, Iran.
Corresponding Author E-mail: Dr.ahmad.vesal@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1081
Abstract
Legonella pneumophila is one of the main pathogenic agents responsible for pneumonia and respiratory tract infections. It has high levels of resistance against commonly used antibiotics. The present investigation was carried out to study the prevalence and antibiotic resistance pattern of L. pneumophila strains isolated from patients suffered from RTIs. Totally, 250 respiratory samples were selected and immediately tested. All samples were cultured and those that were positive for L. pneumophila were subjected to PCR and disk diffusion.Twenty-seven out of 250 respiratory samples (10.80%) were positive for L. pneumophila. Results were also confirmed by lepA gene–based PCR amplification. Prevalence of L. pneumophila in male and female patients were 13.84% and 7.50%, respectively (P < 0.01). Older than 60 years old patients had the highest prevalence of infection with L. pneumophila (P < 0.05). Bacterial strains harbored the highest levels of resistance against ciprofloxacin (81.48%), erythromycin (77.77%), clarithromycin (51.85%) and moxifloxacin (48.14%), while prevalence of resistance against rifampicin (18.51%), doxycycline (22.22%) and azithromycin (25.92%) was low. Primary identification of L. pneumophila positive strains and their regular treatment with rifampicin, doxycycline and azithromycin can reduce the risk of transmission and spread of L. pneumophila.
Keywords
Legonella Pneumophila; Prevalence; Antibiotic resistance pattern; Respiratory tract infection
Download this article as:| Copy the following to cite this article: Rahimi B, Vesal A. Antimicrobial Resistance Properties of Legionella Pneumophila Isolated from the Cases of Lower Respiratory tract Infections. Biomed Pharmacol J 2017;10(1). |
| Copy the following to cite this URL: Rahimi B, Vesal A. Antimicrobial Resistance Properties of Legionella Pneumophila Isolated from the Cases of Lower Respiratory tract Infections. Biomed Pharmacol J 2017;10(1). Available from: http://biomedpharmajournal.org/?p=14158 |
Introduction
Respiratory tract infections (RTIs) are one of the most common and severe types of infectious diseases al-around the world. Documented data revealed that more than 16% of death are occurred due to the RTIs (1,2). RTIs and pneumonia are responsible for more than 50,000 cases in 2010 (1,2). RTIs accounted for about 44,000 hospital admissions with an average length of stay of 6.3 days (1,2). RTIs are usually caused by viruses, however the roles of bacteria are also significant.
Among all bacterial agents which were isolated from the cases of RTIs and pneumonia, Legionella species (Legionella spp.) are one of the most commonly considered pathogens (3-5). Among all species of Legionella, Legionella pneumophila (L. pneumophila) has the highest clinical importance (6-8). It is a causative agents of human legionellosis or Legionnaires Disease (LD) and community-acquired and nosocomial pneumonia (8-10). LD is responsible for more than 18,000 cases of hospitalization in developed countries (10, 11). RTIs and pneumonia caused by L. pneumophila are usually known by confusion, fever, headache, diarrhea, abdominal pain, chills, non-productive cough and myalgia (6-11).
RTIs and pneumonia caused by this bacterium often required antibiotic therapy; However, antibiotic resistant strains of this bacterium cause more sever and dangerous diseases for longer periods of time than susceptible strains (12,13). According to the recent epidemiological studies, L. pneumophila strains show a high prevalence of resistance (50-100%) against commonly used antibiotics including tigecycline, ceftriaxone, rifampicin, azithromycin, erythromycin, moxifloxacin, ciprofloxacin, levofloxacin, doxycycline and clarythromycin (12,13).
According to the uncertain role of L. pneumophila strains as a causative agent of RTIs in male and female of various ages caused us to do this investigation with respect to study the distribution of L. pneumophila in the respiratory samples taken from patients suffered from RTIs as well as study the antimicrobial resistance pattern of bacterial isolates against 10 commonly used antibiotics used for RTIs.
Materials and Methods
Samples Collection and Bacterial Isolation
From January to November 2015, a total of 350 respiratory samples including Broncho Alveolar Lavages (BAL) (n=50) and also respiratory secretions (n=300) were sent to our laboratory center from hospitalized patients suffering from RTIs. In this study, a total of 250 respiratory samples were randomly selected and analyzed for presence of L. pneumophila. At the time of sampling, information about the age, sex and clinical symptoms of the patients were recorded. Ten ml of each sample was immediately transferred to a sterile falcon tube containing ice and was immediately transferred to the laboratory.
Prior to culture, samples were centrifuged for 15 min at 2,500 rpm, and the top 7.5 ml of the resulting suspension was removed. The remaining cell concentrate was mixed and used for culture. Aliquots of 100 µL of prepared samples were spread on duplicate plates of aBCYE selective medium Agar (Difco Laboratories, Detroit, Mich., USA) and to plates containing L-cysteine (0.44mg mL-1), ferric pyrophosphate (0.250 mg mL-1), glycine (3.0 gL-1), vancomycin (0.0025 mg mL-1) and polymyxin B (0.006 mgmL-1), which are named αBCYE-GVP selective agar medium. Plates were incubated at 37ºC in a humidified atmosphere without CO2 during 5 days. Colonies with the typical ground glass appearance of Legionella were sub cultured on two nonselective media, sheep-blood agar and αBCYE agar without L-cysteine. Colonies that grew on αBCYEGVP but not on non-selective media were considered putative Legionella strains, and were Gram stained and subcultured on a selective medium. The identification of putative Legionella strains as L. pneumophila was carried out using Legionella specific latex reagents (Oxoid, Hampshire, England) and direct immunofluorescence assay with poly clonal rabbit sera (m-Tech Alpharetta, Ga., USA).
PCR Confirmation
pneumophila isolates were submitted to DNA extraction using the DNA extraction kit (Fermentas, Germany), according to the manufacturer’s instructions. Set of primers for lepA gene of the L. pneumophila was designed by Khedri et al. (2015) (14). The extracted DNA of each sample was kept frozen at -20°C until used. Primer sequences used for PCR, Legionella-F: 5′- GTTGGGCACTACAGTTATCTCTTC-3′ andLegionella-R: GTTAGTTACTACGGTTTCAATACGAC-3′ (354 bp) were designed from lepA gene of Legionella. PCR reactions were performed in a total volume of 25 µL, including 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 200 µM dNTPs each (Fermentas, Germany), 25 pmoL of each primer, 1.5 U of Taq DNA polymerase (Fermentas, Germany), and 3 µL (40-260 ng/µL) of DNA. The samples were placed in a thermal cycler (Mastercycler gradient, Eppendorf, Germany) with an initial denaturation step at 95°C for 5 min, then amplified for 30 cycles of denaturation at 94°C for 50 s, annealing at 59°C for1 min, extension at 72°C for 1 min and final extension step at 72°C for 5 min. The PCR amplification products (10 μl) were subjected to electrophoresis in a 1% agarose gel in 1X TBE buffer at 80 V for 30 min, stained with ethidium bromide, and images were obtained in a UVIdoc gel documentation system (UK). The PCR products were identified by 100 bp DNA size marker (Fermentas, Germany). A DNA of L. pneumophila ATCC 33152 was used as positive control and DNA of a laboratory isolate strain of E. coli as negative control.
Antibiotic Susceptibility test
pneumophila strains of lower respiratory tract infect ions were cultured on aBCYE selective medium agar (Difco Laboratories, Detroit, Mich., USA). Antimicrobial resistance of the L. pneumophila strains against 10 commonly used antibiotics was determined using the instruction of Clinical and Laboratory Standards Institute guidelines (15). Susceptibility of L. pneumophila isolates were tested against ceftriaxone (30 µg/disk), azithromycin (15 µg/disk), erythromycin (15 µg/disk), ciprofloxacin (5 µg/disk), doxycycline (30 µg/disk), rifampicin (5 µg/disk), tigecycline (15 µg/disk), moxifloxacin (5 µg/disk), clarythromycin (2 µg/disk) and levofloxacin (1 µg/disk) antimicrobial agents (Oxoid, UK). Plates containing the discs were allowed to stand for at least 30 min before incubated at 37ºC in a humidified atmosphere without CO2 during 5 days. The diameter of the zone of inhibition produced by each antimicrobial disc was measured and interpreted using the CLSI zone diameter interpretative standards (15). L. pneumophila ATCC 33152 and S. aureus ATCC 25923 were used as quality control organism in antimicrobial susceptibility determination.
Statistical Analysis
The data were analyzed using SPSS (Statistical Package for the Social Sciences) software and P values were calculated using Chi-square and Fisher’s exact tests to identify statistically significant relationships for the distribution of L. pneumophila and antibiotic resistance between various studied groups of patients. A P value < 0.05 was considered statistically significant.
Results
Table 1 represents the total prevalence of L. pneumophila in the samples taken from patients suffered from RTIs. We found that 27 out of 250 samples (10.80%) were positive for L. pneumophila. Results of the culture method were also confirmed using the lepA gene–based PCR amplification (figure 1). Total prevalence of L. pneumophila in the male and female patients suffered from RTIs were 13.84% and 7.50%, respectively. Statistically significant differences were seen for the prevalence of L. pneumophila between male and female (P < 0.01) and old and young patients (P < 0.05).
Table 1: Total prevalence of Legionella pneumophila in the respiratory samples taken from patients suffered from RTIs.
| Types of samples | No. samples collected | Prevalence of L. pneumophila (%) | PCR confirmation (%) | |
| Male | <20 years | 30 | 2 (6.66) | 2 (6.66) |
| 20-40 years | 32 | 4 (12.50) | 4 (12.50) | |
| 40-60 years | 33 | 5 (15.15) | 5 (15.15) | |
| >60 years | 35 | 7 (20) | 7 (20) | |
| Total | 130 | 18 (13.84) | 18 (13.84) | |
| Female | <20 years | 26 | 1 (3.84) | 1 (3.84) |
| 20-40 years | 31 | 2 (6.45) | 2 (6.45) | |
| 40-60 years | 29 | 2 (6.89) | 2 (6.89) | |
| >60 years | 34 | 4 (11.76) | 4 (11.76) | |
| Total | 120 | 9 (7.50) | 9 (7.50) | |
| Total | 250 | 27 (10.80) | 27 (10.80) | |
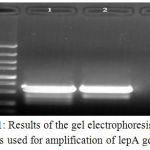 |
Figure 1: Results of the gel electrophoresis of PCR products used for amplification of lepA gene of L.
|
Pneumophila isolated from patients suffered from RTIs. M: 100 bp ladder, 1: Positive sample (354 bp), 2: Positive control and 3: Negative control.
Antibiotic resistance properties of L. pneumophila strains isolated from samples taken from patients suffered from RTIs is shown in table 2. L. pneumophila strains of our investigation harbored the highest levels of resistance against ciprofloxacin (81.48%), erythromycin (77.77%), clarithromycin (51.85%) and moxifloxacin (48.14%). Bacterial strains which were isolated from male patients harbored the higher prevalence of antibiotic resistance than female (P < 0.05). L. pneumophila strains harbored the lowest levels of resistance against rifampicin (18.51%), doxycycline (22.22%) and azithromycin (25.92%).
Table 2: Antibiotic resistance pattern of Legionella pneumophila isolated from the respiratory samples taken from patients suffered from RTIs.
| Samples (No. positive) | Antibiotic resistance pattern (%) | |||||||||
| Cef* | Azi | Ert | Cip | Dox | Rif | Tig | Mox | Clar | Lev | |
| Male (18) | 8 (44.44) | 5 (27.77) | 15 (83.33) | 16 (88.88) | 4 (22.22) | 4 (22.22) | 8 (44.44) | 9 (50) | 10 (55.55) | 8 (44.44) |
| Female (9) | 3 (33.33) | 2 (22.22) | 6 (66.66) | 6 (66.66) | 2 (22.22) | 1 (11.11) | 3 (33.33) | 4 (44.44) | 4 (44.44) | 3 (33.33) |
| Total (27) | 11 (40.74) | 7 (25.92) | 21 (77.77) | 22 (81.48) | 6 (22.22) | 5 (18.51) | 11 (40.74) | 13 (48.14) | 14 (51.85) | 11 (40.74) |
*Cef: ceftriaxone (30 µg/disk), Azi: azithromycin (15 µg/disk), Ert: erythromycin (15 µg/disk), Cip: ciprofloxacin (5 µg/disk), Dox: doxycycline (30 µg/disk), Rif: rifampicin (5 µg/disk), Tig: tigecycline (15 µg/disk), Mox: moxifloxacin (5 µg/disk), Clar: clarythromycin (2 µg/disk), Lev: levofloxacin (1 µg/disk).
Discussion
The results of the present investigation revealed that resistant strains of L. pneumophila had the high ability for presence as a causative agent of the RTIs in Iranian patients. Totally, 10.80% of samples were positive for L. pneumophila which was considerable high. We found that the prevalence of bacteria in male and female patients were 13.84% and 7.50%, respectively. A possible clarification for the higher prevalence of L. pneumophila in male than female is that men usually have more contact with the contaminated external environment. They work outside the house but women usually stay at home and are not in close contact with outside. In fact, most of the Iranian women prefer to work at home. In addition, higher levels of immunity in men than women caused to their infection with resistant strains of L. pneumophila. We also found that older patients had the higher prevalence of bacteria than younger which may be due to their low levels of immunity.
Several investigations were conducted in this field al-around the world. Khedri et al. (2015) (14) reported that of 150 samples tested for presence of L. pneumophila, 18 samples (12%) were positive. They showed that the prevalence of bacteria in male and female patients were 14.40% and 8.30%, respectively. They also showed that older patients had the higher prevalence of L. pneumophila which was completely similar to our findings. Total prevalence of L. pneumophila in the clinical samples of Chaudhry et al. (2000) (16), Yu et al. (2008) (17), Ghotaslou et al. (2013) (18) and Azara et al. (2006) (19) were 5.1%, 13%, 2.85% and 26.66%, respectively. Ngeow et al. (2005) (20) were analyzed 1800 patients for presence of respiratory pathogens. They showed that L. pneumophila is one of the most commonly detected pathogens in studied samples. Similar findings have been reported previously by Nagalingam et al. (2005) (21) and Amemura-Maekawa et al. (2010) (22).
These large differences which were found for the prevalence of L. pneumophila in various researches maybe due to the differences in the type of sample (bronchoalveolar lavage, urine, blood, water, stool, and other clinical samples) tested, number of samples, method of sampling, history of patients (with and without smoking history or other predisposing factors), season of sampling, experimental methodology, geographical area, and climate differences in the areas where the samples were collected, which would have differed between each study.
We found that bacterial strains harbored the highest levels of resistance against ciprofloxacin, erythromycin, clarithromycin and moxifloxacin. These are mainly used for treatment of infections caused by Gram-negative bacteria. Therefore, it showed that treatment of RTIs in Iranian health centers were done according to the results of the disk diffusion and mainly based on the results of Gram staining. The main causes for the high prevalence of resistance against these antibiotics are their irregular, excessive and unauthorized prescription. Several investigations were conducted on the prevalence of antibiotic resistance in L. pneumophila strains of environmental and clinical samples. Moffie and Mouton (1988) (23) reported the low levels of L. pneumophila resistance against rifampicin, erythromycin, norfloxacin and ciprofloxacin. In fact, these antibiotic agents were effective for treatment of RTIs caused by L. pneumophila on 1988 year. Excessive and irregular prescription of these antibiotics caused increase in the levels of resistance such that showed in our results. De Giglio et al. (2015) (13) reported that the levels of minimum inhibitory concentration of azithromycin, ciprofloxacin, levofloxacin, moxifloxacin, and tigecycline were significantly lower than other tested antibiotics. They also showed that doxycycline, tigecycline and cefotaxime are effective antibiotic agents for clinical strains of L. pneumophila. Mallegol et al. (2014) (24) reported similar results for the antibiotic resistance of L. pneumophila strains of clinical samples. High differences which were found in the prevalence of resistance against antibiotics are mainly due to the availability of antibiotics, idea of medical practitioners to prescription of antibiotics, cost of antibiotic agents and also status and conditions exist for prescription of antibiotics.
Conclusions
In conclusion, we identified a large numbers of L. pneumophila in the respiratory samples of male and female patients of various age groups suffered from RTIs as well as their antibiotic resistance pattern. We found that the highest levels of health monitoring should be done for older than 60 years old male patients. We found that judicious and regular prescription of rifampicin, doxycycline and azithromycin can control the risk of RTIs due to the L. pneumophila. We recommended using from simple disk diffusion method to determine proper antibiotic agents for treatment of cases of RTIs due to this bacterium.
Acknowledgement
There is no acknowledgement.
Conflict of Interest
There is no conflict of interest.
References
- Bettering the Evaluation and Care of Health (BEACH) study: URTI / bronchosinusitis in general practice. Australian Institute of Health and Welfare General Practitioner Statistics and Classification Centre, University of Sydney [Westmead Hospital’s Family Medicine Research Centre]. March 2005.
- Hoyert DL, Xu J. Deaths: Preliminary Data for 2011. Natl Vital Stat Rep. 2012; 61(6):1-51.
- Blasi F. Atypical pathogens and respiratory tract infections. Eur Respir J.2004 Jul;24(1):171-81.
CrossRef - Darvishi M, Sadeghi SS. Evaluation of Association of Helicobacter Pylori Infection and Coronary Heart Disease (CHD) among CCU Patients. J PURE APPL MICROBIO. 2016;10(4):2621-2626.
CrossRef - Chaudhry R,Valavane A, Mohan A, Dey AB. Legionella pneumophila infection associated with renal failure causing fatality in a known case of sarcoidosis. Indian J Med Microbiol. 2014 Jul-Sep;32(3):324-7.
CrossRef - Furugen M,Higa F, Hibiya K, Teruya H, Akamine M, Haranaga S, Yara S, Koide M, Tateyama M, Mori N, Fujita J. Legionella pneumophila infection induces programmed cell death, caspase activation, and release of high-mobility group box 1 protein in A549 alveolar epithelial cells: inhibition by methyl prednisolone. Respir Res. 2008 May 1;9:39.
CrossRef - Gobin I, Newton PR, Hartland EL, Newton HJ. Infections caused by non pneumophila species of Legionella. Rev Med Microbiol. 2009; 20(1):1-11.
CrossRef - Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002; 15(3):506-26.
CrossRef - Newton HJ, Ang DK, van Driel IR, Hartland EL. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev. 2010; 23(2):274-98.
CrossRef - Lück C. Legionella: a case for culture. Indian J Med Res. 2010; 131(6):736-8.
- Diederen BM, de Jong CM, Marmouk F, Kluytmans JA, Peeters MF, Van der Zee A. Evaluation of real-time PCR for the early detection of Legionella pneumophila DNA in serum samples. J Med Microbiol. 2007; 56(Pt 1):94-101.
CrossRef - Nielsen K,Bangsborg JM, Høiby N. Susceptibility of Legionella species to five antibiotics and development of resistance by exposure to erythromycin, ciprofloxacin, and rifampicin. Diagn Microbiol Infect Dis. 2000 Jan;36(1):43-8.
CrossRef - De Giglio O,Napoli C, Lovero G, Diella G, Rutigliano S, Caggiano G, Montagna MT. Antibiotic susceptibility of Legionella pneumophila strains isolated from hospital water systems in Southern Italy. Environ Res. 2015 Oct;142:586-90.
CrossRef - Khedri F, Alaei Faradonbeh F, Eliyasi M, Barghi A, Doosti A, Emad P, Alaei Faradonbeh A. Molecular depiction of lepa, lida, ralf, rtxa and lvhb virulence factors of Legionella Pneumophila isolated from respiratory tract infections. InternatIonal archIves of MedIcIne 2015; 8: 1-11.
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard-Ninth Edition (M2-A9). United States: Clinical and Laboratory Standards Institute; 2012.
- Chaudhry R, Dhawan B, Dey AB. The incidence of Legionella pneumophila: a prospective study in a tertiary care hospital in India. Trop Doct. 2000; 30(4):197-200.
CrossRef - Yu PY, Lin YE, Lin WR, Shih HY, Chuang YC, Ben RJ, et al. The high prevalence of Legionella pneumophila contamination in hospital potable water systems in Taiwan: implications for hospital infection control in Asia. Int J Infect Dis. 2008; 12(4):416-20.
CrossRef - Ghotaslou R, Yeganeh Sefidan F, Akhi MT, Soroush MH, Hejazi MS. Detection of Legionella Contamination in Tabriz Hospitals by PCR Assay. Adv Pharm Bull. 2013; 3(1):131-4.
- Azara A, Piana A, Sotgiu G, Dettori M, Grazia Deriu M, Masia MD, et al. Prevalence study of Legionella spp. contamination in ferries and cruise ships. BMC Publ Health. 2006; 6:100.
CrossRef - Ngeow YF, Suwanjutha S, Chantarojanasriri T, Wang F, Saniel M, Alejandria M, Hsueh PR, Ping-Ing L, Park SC, Sohn JW, Aziah AM, Liu Y, Seto WH, Ngan CC, Hadiarto M, Hood A, Cheong YM. An Asian study on the prevalence of atypical respiratory pathogens in community-acquired pneumonia. Int J Infect Dis 2005, 9: 144-153.
CrossRef - Nagalingam NA, Adesiyun AA, Swanston WH, Bartholomew M. Seroprevalence of Legionella pneumophila in Pneumonia Patients in Four Major Hospitals in Trinidad and Tobago. West Indian Med J 2005, 54: 375-378.
CrossRef - Amemura-Maekawa J, Kura F, Helbig JH, Chang B, Kaneko A, Watanabe Y, Isobe J, Nukina M, Nakajima H, Kawano K,Tada Y, Watanabe H. Working Group for Legionella in Japan. Characterization of Legionella pneumophila isolates from patients in Japan according to serogroups, monoclonal antibody subgroups and sequence types. J Med Microbiol 2010, 59: 653659.
CrossRef - Moffie BG,Mouton RP. Sensitivity and resistance of Legionella pneumophila to some antibiotics and combinations of antibiotics. J Antimicrob Chemother. 1988 Oct;22(4):457-62.
CrossRef - Mallegol J, Fernandes P, Melano RG, Guyard C. Antimicrobial activity of solithromycin against clinical isolates of Legionella pneumophila serogroup 1. Antimicrob Agents Chemother. 2014;58(2):909-15.
CrossRef








