Chitralekha Saikumar1 , Punithavathi Velmurugan2
, Punithavathi Velmurugan2 and Aishwarya Jothi Ramalingam3*
and Aishwarya Jothi Ramalingam3*
Department of Microbiology, Sree Balaji Medical College and Hospital, Bharath Institute of Higher Education and Research, Chennai, Tamil Nadu, India.
Corresponding Author E-mail:jhaish87@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/3027
Abstract
Millions of people worldwide suffer from chronic suppurative otitis media (CSOM), a recurrent infection of the ear that is particularly common in low-resource environments. Although there are several etiological factors, bacterial infection is a significant one. The two most frequently isolated bacteria are Staphylococcus aureus and Pseudomonas aeruginosa. Due to the emergence and spread of bacterial resistance to several antimicrobial agents and also their ability to form biofilms, treatment and recovery are made more challenging. Our aim was to identify the clinico-bacteriological profile, their antimicrobial susceptibility pattern and a molecular method to identify the antibiotic resistance genes of the most common isolates in this study. Following approval from the Institutional Human Ethics Committee (IHEC) and patient informed consent, we obtained ear discharge samples from 100 CSOM patients over a two-year period. The isolates were recognized by conventional microbiological methods. Polymerase chain reaction (PCR) was used for molecular characterisation in order to identify genes linked to antibiotic resistance (e.g., efflux pump-related genes associated with P. aeruginosa and mecA for S. aureus). Of the 100 patient samples, 84 samples showed positive results for culture; 58% of the Gram-positive cocci were S. aureus and 42% of Gram-negative bacilli were P. aeruginosa. Methicillin resistance in S. aureus is indicated by the presence of the mecA gene (43%) and carbapenem resistance in P. aeruginosa is indicated by the presence of the blaVIM gene (22%), according to genotypic study. Our work provides insight with regard to the genetic makeup of P. aeruginosa and S. aureus in CSOM patients, which demonstrates that these bacteria have several resistance genes that enable them to withstand antimicrobial therapy.
Keywords
Chronic suppurative otitis media (CSOM); Coagulase negative Staphylococcus (CoNS); multidrug resistant (MDR); Methicillin resistant Staphylococcus aureus (MRSA); Pseudomonas aeruginosa; Staphylococcus aureus
Download this article as:| Copy the following to cite this article: Saikumar C, Velmurugan P, Ramalingam A. J. Bacteriological Profile, Drug Resistance Pattern and its Molecular Characterisation Among Patients with Chronic Suppurative Otitis Media at a Tertiary Care Hospital in South India. Biomed Pharmacol J 2024;17(4). |
| Copy the following to cite this URL: Saikumar C, Velmurugan P, Ramalingam A. J. Bacteriological Profile, Drug Resistance Pattern and its Molecular Characterisation Among Patients with Chronic Suppurative Otitis Media at a Tertiary Care Hospital in South India. Biomed Pharmacol J 2024;17(4). Available from: https://bit.ly/4imfqLc |
Introduction
Chronic suppurative otitis media (CSOM) is characterised by a persistent infection of the middle ear cleft, which includes the middle ear, mastoid, eustachian tube, as well as a perforated tympanic membrane and otorrhea, that lasts for two weeks or more. It is sometimes referred to as chronic tympanomastoiditis, chronic otomastoiditis, and chronic active mucosal otitis media.1 This typically affects children and develops as a side effect of acute otitis media.2 In about half of the patients, suppurative discharge and hearing loss are the most prevalent presentations.3 It is more prevalent in developing countries, especially among lower socioeconomic classes with limited health care, overcrowding, poor hygiene, and malnutrition which significantly lowers one’s quality of life.4,5 Risk factors include prior upper respiratory tract infections, traumatic tympanic rupture, and recurrent episodes of acute otitis media.6
Globally, the CSOM prevalence ranges from 1 to 46%. It is more prevalent in South-east Asian countries, Africa, the Western Pacific.7 CSOM is uncommon in the United states, Australia, Middle East and Europe8. It is estimated that there are between 65 and 300 million instances globally, and that 60% of these cases result in substantial hearing impairment.9 It has been stated that CSOM problems result in 28,000 fatalities annually worldwide. The annual cost of otitis media and its consequences, such as CSOM10, is estimated to be approximately $5 billion in the United States. Anaerobes, aerobes, and fungi are considered potential pathogens in CSOM, though their reported profiles and frequencies vary depending on the geography, age, and presence of complications such as cholesteatoma.11 Aerobic bacteria such as Escherichia coli, Proteus species, Klebsiella pneumoniae and Streptococcus pyogenes are commonly isolated after Pseudomonas aeruginosa and Staphylococcus aureus, while Bacteroides, Peptostreptococcus and Propionibacterium are the most common anaerobes. As with other bacterial infections, multidrug-resistant (MDR) bacteria are a growing concern in CSOM.12 A significant incidence of multidrug resistance against amoxycillin, amoxycillin/clavulanic acid, ampicillin, cephalosporins, macrolides, co-trimoxazole, quinolones has resulted in adverse treatment outcomes in recent years.13
In addition to preventing complications from CSOM, early identification of the causative bacteria and their susceptibility is essential for a satisfactory clinical recovery. Appropriate antimicrobial therapy is employed to eliminate the bacterial agents responsible for otitis media; nevertheless, the majority of microorganisms are developing resistance to antibiotics.14
The bacteria causing CSOM are becoming more MDR as a result of antibiotic overuse and misuse causing this infection to spread quickly in underdeveloped nations15, which necessitates routine surveillance of the microbiological and susceptibility profiles.16 Additionally, knowing the range of bacteria and how susceptible they are to antibiotics will help us understand CSOM better and will be essential for successful empirical treatment.14 Accordingly, the current study identified the clinico-bacteriological profile of CSOM, examined their susceptibility pattern of different antibiotics and used a molecular technique to identify the antibiotic resistance genes of the most common isolates.
Materials and Methods
Study Area
The study consisted of 100 CSOM patients attending ENT department at a tertiary care facility in Chennai, South India. The ear discharge samples were collected using sterile swabs and transported to the Department of Microbiology, Central Laboratory.
Inclusion criteria: a) 100 clinically diagnosed CSOM patients of different age groups. b) active ear discharge for at least 3 months c) discharge from one/both ears were included in the study.
Exclusion criteria: a) active ear discharge of less than 3 months duration b) patients who were recently treated for CSOM c) patients with ear discharge and intact tympanic membrane d) samples which had growth other than bacteria e) patients with congenital ear, obstructed middle ear, malignancy, prior ear surgery were excluded from the study.
Study Design
A cross-sectional study was conducted from August 2022 to July 2024 after obtaining approval from Institutional Human Ethics Committee (IHEC) (Ref. no. 002/SBMCH/IHEC/2022/1806) and informed consent from all the participants. The samples were subjected to microbiological analysis and genotypic detection of the predominant bacteria by PCR.
Microbiological Analysis
Two ear swabs from each patient were aseptically taken after a thorough inspection of the ear using an otoscope. One ear swab was subjected to direct microscopy and the other was used to inoculate on nutrient agar, MacConkey agar and blood agar. The plates were incubated at 37℃ for 18-24 hrs and identification of organisms was done based on standard bacteriological techniques.17 Muller-Hinton agar was used for antimicrobial susceptibility testing, and the results were interpreted in accordance with Clinical and Laboratory Standards Institute (CLSI) recommendations 2022.18
Antibiotics Tested for Staphylococci
Penicillin (10μg), cefoxitin (30μg), ciprofloxacin (5μg), levofloxacin (5μg), gentamicin (10μg), erythromycin (15μg), clindamycin (2μg), linezolid (15μg), tetracycline (30μg), co-trimoxazole (25μg), vancomycin (30μg), teicoplanin (30μg)
Antibiotics Tested for other Gram Positive Cocci
Penicillin (10μg), ampicillin (10μg), chloramphenicol (30μg), erythromycin (15μg), clindamycin (2μg), tetracycline (30μg), levofloxacin (5μg), high level gentamicin (120μg), linezolid (15μg), vancomycin (30μg), cefepime (30μg), cefotaxime (30μg), ceftriaxone (30μg)
Antibiotics Tested for Pseudomonas Species
Amikacin (30μg), gentamicin (10μg), ciprofloxacin (5μg), levofloxacin (5μg), piperacillin-tazobactam (100/10μg), ceftazidime (30μg), cefepime (30μg), aztreonam (30μg), meropenem (10μg), cefoperazone-sulbactam (75/30μg)
Antibiotics Tested for other Gram Negative Bacilli
Ampicillin (10μg), amoxycillin-clavulanic acid (30μg), levofloxacin (5μg), amikacin (30μg), gentamicin (10μg), cefepime (30μg), ceftriaxone (30μg), cefotaxime (30μg), ceftazidime (30μg), cefazolin (30μg), piperacillin-tazobactam (100/10μg), meropenem (10μg), co-trimoxazole (25μg), aztreonam(30μg)
Organisms resistant to more than 3 class of antibiotics was considered as multidrug resistant (MDR). Phenotypic tests for extended spectrum beta-lactamase (ESBL) & methicillin-resistant Staphylococci were performed18. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were utilized as controls. The media, reagents, antibiotic discs, ATCC strains were purchased from HiMedia, Mumbai, India. For additional validation of the pathogenic organism and its antimicrobial susceptibility, the Vitek-2 compact automated system (Vitek-2 GN card and Vitek-2 GP card, BioMérieux Inc., Durham, NC) was utilized. Polymerase chain reaction (PCR) was done to find resistant genes of predominant bacteria.
Genotypic detection of predominant bacteria19,20
PCR amplification was done for all the MDR Pseudomonas and methicillin resistant S. aureus isolates to detect the genes encoding resistance blaVIM genes and mecA genes respectively. Individual colonies were picked up from the quadrant streak plates and were amplified by 16S rRNA primer. A PCR template was created by dissolving each colony in 23.8 µl of distilled water. Every colony underwent PCR, and the amplified PCR products were examined in a 1% agarose gel to ensure their quality and quantity. Primer details: blaVIM F:GATGGTGTTTGGTCGCATA R: CGAATGCGCAGCACCAG mecA F-AAAATCGATGGTAAAGGTTGGC) R: AGTTCTGCAGTACCGGATTTTGC3′)
Statistical Analysis
For data entry, Microsoft Excel was used. The data were analysed and P-value was calculated using IBM Corp.’s Armonk, NY SPSS, Version 25.0. Tables showed the frequency and percentage representations of the descriptive statistics.
Results
In our study, out of 100 CSOM patient samples, 59% were from male patients and 41% were from female patients. Table 1 shows the distribution of samples among various age groups.
Table 1: Distribution of samples among various age groups
|
Age Group |
Frequency (n) |
% |
|
<10 years |
18 |
18 |
|
11-20 |
19 |
19 |
|
20-40 |
26 |
26 |
|
40-60 |
27 |
27 |
|
>60 |
10 |
10 |
|
Total |
100 |
100 |
|
Mean age |
44.8 years |
|
Unilateral involvement of the ear was seen in 95% of the patients (right ear-56 and left ear-39) and bilateral involvement was seen in 5% of the patients. Out of the 100 CSOM patients, 84 patients yielded growth in culture. Among the culture positive patients, 17 (20%) had polymicrobial growth and 67 (80%) had monomicrobial growth which yielded a total of 101 isolates. Out of 101 isolates, 53 (52%) were Gram-negative bacilli and 48 (48%) were Gram-positive cocci. Table 2 and table 3 shows the prevalence of Gram-negative bacilli and Gram-positive cocci among CSOM patients, respectively.
Table 2: Prevalence of Gram-negative bacilli among CSOM patients
|
GNB isolates |
Frequency |
|
Pseudomonas aeruginosa |
22 (42) |
|
Klebsiella pneumoniae |
13 (25) |
|
E.coli |
10 (19) |
|
Proteus vulgaris |
5 (9) |
|
Proteus mirabilis |
2 (4) |
|
Enterobacter species |
1 (2) |
|
Total |
53 (100) |
Table 3: Prevalence of Gram-positive cocci among CSOM patients
|
GPC isolates |
Frequency |
|
Staphylococcus aureus |
28 (58) |
|
CoNS |
14 (29) |
|
Streptococcus pyogenes |
2 (4) |
|
S. pneumoniae |
1 (2) |
|
other Streptococcus spp. |
1 (2) |
|
Enterococcus faecalis |
2 (4) |
|
Total |
48 (100) |
The prevalence of antimicrobial resistance to the most commonly used antibiotics are depicted in figures 1 and 2.
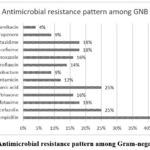 |
Figure 1: Antimicrobial resistance pattern among Gram-negative bacilli |
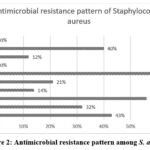 |
Figure 2: Antimicrobial resistance pattern among S. aureus |
It was found that 32% of P. aeruginosa, 46% of K. pneumoniae, 40% of E. coli, 20% of P. vulgaris were extended spectrum beta-lactamases (ESBL) producers. The prevalence of methicillin resistant S. aureus (MRSA) and methicillin resistant CoNS (MRCoNS) by phenotypic detection method were found to be 43% and 57% respectively.
The prevalence of blaVIM gene among MDR P. aeruginosa isolated from CSOM patients by PCR is shown in table 4.
Table 4: PCR identification of blaVIM gene among MDR P. aeruginosa
|
blaVIM |
No. of MDR P. aeruginosa isolates (%) |
|
Positive |
2 (22%) |
|
Negative |
7 (78%) |
|
Total |
9 (100%) |
Figure 3 shows the amplification of blaVIM gene by PCR in 2 selected P.aeruginosa isolates
 |
Figure 3: L1 – 1000Bp ladder, L2 – negative control for 710bp blaVIM L3 to L4 – blaVIM amplification positive strains. |
The table 5 describes the prevalence of mecA gene among Staphylococci isolated from CSOM patients.
Table 5: Prevalence of mecA gene among Staphylococci
|
Staphylococci |
Total (n=20) |
mecA positive |
mecA negative |
P Value |
OR |
95% CI
|
|
MRSA |
12 |
12 (100%) |
0 |
0.068 |
1.333 |
0.89-1.98 |
|
MRCoNS |
8 |
6 (75%) |
2 (25%) |
Figure 4 shows the amplification of mecA gene by PCR in 5 selected S.aureus isolates
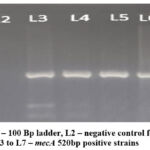 |
Figure 4: L1 – 100 Bp ladder, L2 – negative control for mecA, L3 to L7 – mecA 520bp positive strains |
Discussion
CSOM is a chronic disease that affects people of all ages and has a low recovery rate. The prevalence of ear infections varies among different populations. The WHO reports that the prevalence of CSOM in India is 7.8%, placing the country among the highest in the world and necessitating prompt attention to address a serious public health issue.5 In order to prevent major complications from CSOM, early microbiological identification is essential for both timely and efficient treatment.
The current study reported male and female prevalence were 59% and 41% respectively. A recent study reported that about 58% infected were males, 42% were females.21 A number of studies have revealed a male predominance, which may be related to their active lifestyles and hobbies like diving and swimming.22 Recurrent infections and intermittent ear discharge can result from swimming-induced middle ear soiling. The increase in health awareness among women may be the reason for their lesser preponderance, since they tend to seek care earlier.
In our study, the predominant age was 40-60 years (27%) and 20-40 years (26%) followed by 11-20 years (19%), <10 years (18%) and >60 years (10%). A similar study reported 40% infected were children and 60% were adults.23 Another recent study found that the predominant age group was 41-65 years (54%).22 The fact that eustachian tubes in children are shorter, thinner, and more horizontal than those of adults may be the cause of the high occurrence of CSOM in children.24 Additionally, there is an increased risk of upper respiratory tract infections (URTIs), which can lead to ear infections, in this age range.
The current study reported culture positivity for 84% samples. Among them 17% were polymicrobial and 67% were monomicrobial. A range of 84% to 91.18% has been recorded in several Indian researches on the culture positivity rate. A recent study reported 73% of the samples were positive for culture.25 Another study reported higher culture positive reports (94%).26 In contrast, a study reported only 69.2% culture positive.24 Studies reported with less percentage of culture positive may be due to patient’s prior treatment with broad spectrum antibiotics before obtaining the sample.
Among 48 Gram positive cocci isolates, predominant species was S. aureus (58%) followed by CoNS (29%), Streptococcus spp. (8%), Enterococcus spp. (4%). Out of 53 GNB, P. aeruginosa (42%) was the predominant bacilli followed by K. pneumoniae (25%), E. coli (19%), P. vulgaris (9%), P. mirabilis (4%) and Enterobacter spp. (2%). According to a research, S. aureus (30.35%)27 and P. aeruginosa (44.64%) were the two most common bacteria which was consistent with our study. Predominance of Gram-negative bacilli (59.74%) was reported in a related study where P. aeruginosa had the highest incidence (45.5%), followed by S. aureus (37.7%).28 A higher rate of P. aeruginosa has its own consequences because Pseudomonas is a major pathogen in nosocomial infection and can spread resistance-carrying plasmid to other species. According to another study, S. aureus was predominant followed by Pseudomonas species.6 S. aureus is a common cause of middle ear infections, and its high resistance strain carriage in the upper respiratory tract and external auditory canal can be ascribed to this fact. Pseudomonas species are primarily considered as secondary invaders from the external auditory canal that enter the middle ear through a tympanic membrane defect brought on by an acute episode of otitis media. When the resistance is low, bacteria like Proteus spp., Klebsiella spp. and E. coli turn into opportunistic pathogens in the middle ear.
In this study, among S. aureus alarming resistance to ciprofloxacin (61%) was observed, which may have resulted from irrational use, insufficient dosage, or over-the-counter availability. On the other hand, the lesser resistance to levofloxacin (12%) was seen. MRSA which is 100% susceptible to vancomycin and linezolid was identified which was in line with a similar study.6 Gram-negative bacteria in our investigation were more susceptible to amikacin, piperacillin-tazobactam and meropenem. Maximum resistance to ampicillin and cephalosporins were observed. These concur with the similar research conducted in CSOM patients.29
In the present study we reported among 12 MRSA, all the isolates showed positive for mecA gene amplification and among 8 CoNS isolated 6 (75%) were positive for mecA gene. In a study among MRSA isolates from ear swab, 50% of the isolates recovered mecA gene and 66.6% of the MRCoNS were positive for mecA gene.30 Among the predominant Gram-negative bacilli in the current study, blaVIM gene by PCR amplification of MDR Pseudomonas strains were reported. Among 9 isolates of MDR P. aeruginosa, 2(22%) were positive for blaVIM gene. Research has revealed that MDR P. aeruginosa clones are linked to metallobeta-lactamases (MBL) genes, primarily VIM and IMP, which can be acquired through horizontal gene transfer or chromosomal mutations.31 Another study reported 40% carbapenem resistant P. aeruginosa (CRPA) isolates harbored the blaVIM genes respectively.32 In contrast to our research, a study reported that 39% of the isolates of P. aeruginosa were MDR and that no isolate had blaVIM genes found in it.33 The aforementioned research shown that the microbial and their genotypic profile varies depending on patient population and geographical distribution between different places. Therefore, it strongly suggests that the microbiological profile in every geographic area be regularly examined and updated.
Limitations of this Study
In our study, on comparing the prevalence of mecA gene among Staphylococci, the p-value was 0.068 (nearly statistically significant) because of the less number of MRSA and MRCoNS isolated. Also, other causes of CSOM such as anaerobes, fungi were not included in this study.
Hence, further research is encouraged to get beyond these limitations.
Conclusion
In this study we determined that S. aureus and P. aeruginosa were the primary etiological agents of CSOM. Understanding the bacterial profile and their antimicrobial susceptibility pattern are crucial for patient therapy as it determines when to start antimicrobial therapy, which lowers treatment costs. The strains of P. aeruginosa that produced blaVIM were the predominant ones in our study. Threatening therapeutic options, the advent of carbapenem resistance highlights the necessity for continued epidemiology and antimicrobial susceptibility research as well as long-term monitoring of antibiotic prescription practices. Hospital infection control teams should be concerned about the ongoing existence of these harmful organisms and how they may spread both within and between hospitals. The most successful way to treat CSOM is to choose topical or systemic antibiotics carefully, based on the culture and sensitivity reports, while maintaining dry ears to avoid drug resistance, unintended antibiotic delivery, and other problems. Antibiotic therapy is not always necessary for otitis media, but it should be used with caution when prescribing medication in cases of middle ear infection to avoid the development and dissemination of resistant bacterial strains in hospital and community settings.
Acknowledgement
The authors would like to thank the Central Diagnostic Laboratory, the Central Research Laboratory, Sree Balaji Medical College and Hospital (SBMCH) for providing the laboratory facilities and the Department of ENT for their support.
Funding Sources
The authors received no financial support for the research, authorship, and/or publication of this article
Conflicts of Interest
The authors do not have any conflict of interest
Data Availability Statement
The manuscript incorporates all datasets produced or examined throughout this research study.
Ethics Statement
This cross-sectional study was conducted after obtaining approval from Institutional Human Ethics Committee (IHEC), Sree Balaji Medical College and Hospital (SBMCH), Bharath Institute of Higher Education and Research (BIHER), Chennai-600044 (Ref. no. 002/SBMCH/IHEC/2022/1806)
Informed Consent Statement
Informed consent was obtained from all the participants in the study and it conforms to the standards currently applied in our country. The privacy rights of human subjects has been observed in our study.
Author Contributions
Each author mentioned has significantly and directly contributed intellectually to the project and has given their approval for its publication.
Chitralekha Saikumar: Visualization, Resources, Supervision, Project Administration.
Punithavathi Velmurugan: Data Collection, Analysis, Writing – Original Draft
Aishwarya J Ramalingam: Conceptualization, Methodology, Writing –Review & Editing.
References
- Morris P. Chronic suppurative otitis media. BMJ Clin Evid. 2012;2012:0507.
- Mahajan T, Dass A, Gupta N, Chander J, Saini V, Pol SA. Bacteriological Profile in Attico-antral type of Chronic Suppurative Otitis Media. Indian J Otolaryngol Head Neck Surg. 2019;71(Suppl 2):1412-1421
CrossRef - Rangaiah ST, Dudda R, Prasad MH, Balaji NK, Sumangala B, Gudikote MM. Bacteriological profile of chronic suppurative otitis media in a tertiary care hospital. Int J Otorhinolaryngol Head Neck Surg. 2017;3:601-5
CrossRef - Sah BP, Chettri ST, Bhattarai NR, Shah SP, Paudel D, Sarraf DP, Mishra S. Microbiological profile and their antibiotic sensitivity pattern in patients of chronicsuppurative otitis media at eastern tertiary care center of Nepal. IP J Otorhinolaryngol Allied Sci. 2020;3(3):86-90
CrossRef - Sharma K, Oberoi L, Narula V. Present scenario of microbiological pattern in chronic suppurative otitis media and its management guidelines. J Acad Clin Microbiol. 2017;19:47-53
CrossRef - Smitha N R, Jnaneshwara K B, Patil A B, Harshika Y K, Medegar S. A study of aerobic bacteriological profile of chronic suppurative otitis media in a tertiary care hospital, South India. Indian J Microbiol Res. 2018;5(4):470-47
CrossRef - Acuin J. Chronic suppurative otitis media. BMJ Clin Evid. 2007;2007:0507.
CrossRef - Vikram BK, Khaja N, Udayashankar SG, Venkatesha BK, Manjunath D. Clinico-epidemiological study of complicated and uncomplicated chronic suppurative otitis media. J Laryngol Otol. 2008;122(5):442-446.
CrossRef - Shrestha B, Amathya R, Shrestha I, Ghosh I. Microbiological profile of chronic suppurative otitis media. Nepalese J ENT Head and Neck Surg. 2011;2(2):6-7.
CrossRef - Agrawal R, Khatri P, Parihar R, Shah H. Microbial assessment of chronic suppurative otitis media in a tertiary care center of Rajasthan. Int J Health Sci Res. 2017;7:120-6.
- Malkappa SK, Kondapaneni S, Surpam RB, Chakraverti TK. Study of aerobic bacterial isolates and their antibiotic susceptibility pattern in chronic suppurative otitis media. Indian J of Otol. 2012;18:136-9.
CrossRef - Kumar H, Seth S. Bacterial and fungal study of 100 cases of chronic suppurative otitis media. J Clin Diagn Res 2011;5:1224-7.
- Shariff ME. Analysis of hearing loss by pure tone audiometry in patients with chronic suppurative otitis media. Natl J Physiol Pharm Pharmacol. 2019;9:515-8.
CrossRef - Srivastava A, Singh RK, Varshney S, Gupta P, Bist SS,Bhagat S, Gupta, N. Microbiological evaluation of an active tubotympanic type of chronic suppurative otitis media. Nepalese J ENT Head Neck Surg. 2010;1:14-6
CrossRef - Shaheen MM, Raquib A, Ahmad SM. Chronic suppurative otitis media and its association with socio-econonic factors among rural primary school children of Bangladesh. Indian J Otolaryngol Head Neck Surg. 2012;64:36-41.
CrossRef - Khatoon A, Rizvi M, Sultan A, Khan F, Sharma M, Shukla I, Khan HM. Chronic suppurative otitis media: A clinico-microbiological menace. Int J Res Med Sci. 2015;3:1932-6.
CrossRef - Patricia MT: Bailey & Scott’s Diagnostic Microbiology. Elsevier, St. Louis, MO; 2014.
- Clinical and Laboratory Standards Institute (CLSI): Performance Standards for Antimicrobial Susceptibility Testing. CLSI Document M100, 32nd edition. Clinical and Laboratory Standards Institute, Wayne, PA; 2022.
- Fallah F, Borhan RS, Hashemi A. Detection of bla(IMP) and bla(VIM) metallo-β-lactamases genes among Pseudomonas aeruginosa strains. Int J Burns Trauma. 2013;3(2):122-124.
- Pournajaf A, Ardebili A, Goudarzi L, Khodabandeh M, Narimani T, Abbaszadeh H. PCR-based identification of methicillin-resistant Staphylococcus aureus strains and their antibiotic resistance profiles. Asian Pac J Trop Biomed. 2014;4(Suppl 1):S293-S297.
CrossRef - Elyounsi N, Said A, Abuhelala H, Alsharif H, Elkammoshi A. Isolation and Identification of the Bacteria that Causes Otitis Media in Medical Center Hospitals Tripoli, Libya. Alq J Med App Sci. 2023;6(2):666-671.
- Appiah-Korang L, Asare-Gyasi S, Yawson AE, Searyoh K. Aetiological agents of ear discharge: a two year review in a teaching hospital in Ghana. Ghana Med J. 2014;48(2):91-95.
CrossRef - Suryani S, Dharma A, Nasir N. Isolation and identification of pathogenic bacteria secretion of chronic suppurative otitis media patients. Rasayan J. Chem. 2018 1;11(3):1139-43.
CrossRef - Wan Draman WNA, Md Daud MK, Mohamad H, Hassan SA, Abd Rahman N. Evaluation of the current bacteriological profile and antibiotic sensitivity pattern in chronic suppurative otitis media. Laryngoscope Investig Otolaryngol. 2021;6(6):1300-1306.
CrossRef - Jobin S R, J W Prakash, Jagatheeswary PAT, Rema Devi, Subitha S, L Suresh Babu, Kiran Gopal, Bewin Oral J, P S Aanie. Bacterial profile and antimicrobial susceptibility patterns of otitis media in health care centre, southern Trivandrum, Kerala, India. MedPulse International Journal of Microbiology. 2020;14(1):01-05.
CrossRef - Dhakal A, Sharma S. Microbiological Spectrum Causing Chronic Suppurative Otitis Media and Determination of the Antibiotic Sensitivity Pattern of Isolated Bacteria. Medical Journal of Eastern Nepal. 2023;2(02):16-20.
CrossRef - Kashyap S, Pandey A, Thakuria B, Saxena AK, Asthana AK,Madan M. Resistant microorganisms isolated from cases of chronic suppurative otitis media: A therapeutic concern. Natl. lab. med.2017;6:M001-6.
- Kumar H, Seth S. Bacterial and fungal study of 100 cases of chronic suppurative otitis media. J Clin Diagn Res. 2011;5(6):1224-7.
- Harshika YK, Sangeetha S, Prakash R. Microbiological profile of CSOM and their antibiotic sensitivity pattern in a tertiary care hospital. Int J Curr Microbiol App Sci. 2015;4(12):735-743.
- Ibadin EE, Enabulele IO, Muinah F. Prevalence of mecA gene among staphylococci from clinical samples of a tertiary hospital in Benin City, Nigeria. Afr Health Sci. 2017;17(4):1000-1010.
CrossRef - Umar JB, Ibrahim MM, Tom IM, Umoru AM, Isa T. Pseudomonas aeruginosa in otitis media. Int J Med. 2016;4:55.
CrossRef - Dogonchi AA, Ghaemi EA, Ardebili A, Yazdansetad S, Pournajaf A. Metallo-β-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: A potential threat to clinical therapeutics. Ci Ji Yi Xue Za Zhi. 2018;30(2):90-96.
CrossRef - ALjaafreha LY, Tawalbeh M, Shehabi AA. Otitis external infections among Jordanian patients with emphasis on pathogenic characteristics of Pseudomonas aeruginosa isolates. Open Microbiol J. 2019;13:292-296.
CrossRef








