Soumya Satpathy 1 , Sanat Kumar Bhuyan 2
, Sanat Kumar Bhuyan 2 and Ruchi Bhuyan1*
and Ruchi Bhuyan1* ,
,
1Department of Medical Research, IMS and SUM Hospital, Siksha O Anusandhan Deemed to be University,
2Institute of Dental Sciences, SOA Deemed to be University, Bhubaneswar, Odisha, India.
Corresponding Author E-mail: ruchibhuyan@soa.ac.in
DOI : https://dx.doi.org/10.13005/bpj/2890
Abstract
Oral cancer was recognized as the most common type of cancer in South Asian countries including India. As concurrent chemoradiotherapy leads to various associated new problems, there is always a need for improved therapies without side effects. Natural plant products used since ancient times may fill the gap. Phytoconstituents can activate various cell death pathways, such as apoptosis, autophagy, or pyroptosis to treat oral tumors. Numerous studies have already been done to date to enlighten the detailed mechanism of the use of phytoconstituents in these cell-signaling pathways. As the majority of the studies emphasized the apoptotic pathway, the least reports are found on autophagy. ‘AMPK’ and ‘mTOR’ have been acknowledged to be the key signaling compounds that modulate autophagy. Therefore the objective of this article is to discuss the mechanism of autophagy concerning phytoconstituents in the treatment of oral carcinoma.
Keywords
AMPK; Autophagy; mTOR; Oral cancer; Phytoconstituents
Download this article as:| Copy the following to cite this article: Satpathy S, Bhuyan S. K, Bhuyan R. Targeting Autophagic Pathway in Oral Cancer Therapy Through Phytoconstituents: A Short Review. Biomed Pharmacol J 2024;17(2). |
| Copy the following to cite this URL: Satpathy S, Bhuyan S. K, Bhuyan R. Targeting Autophagic Pathway in Oral Cancer Therapy Through Phytoconstituents: A Short Review. Biomed Pharmacol J 2024;17(2). Available from: https://bit.ly/3V8ro0l |
Introduction
The mechanism of autophagy is a cellular recycling process that attempts to maintain the removal of unwanted proteins as well as unhealthy or old organelles1. The molecular mechanics of the process of autophagy through various types of actions are least specified2. There are several chemotherapy-resistant mechanisms available which include cell death processes like autophagy and apoptosis, multi-drug resistance, cancer cell heterogeneity, and cancer micro-environment pressure-induced genetic or epigenetic modifications. Along with the above factors, alterations in two self-disparaging procedures(apoptosis and autophagy) might initiate better therapies3,4 for cancer treatment. Phytochemicals trigger different cell death pathways, such as apoptosis, autophagy, or pyroptosis.
Numerous studies have already been performed to date to explain the detailed mechanism of the use of phytochemicals in these cell-signaling pathways. The potential purpose of phytochemicals during the process of apoptosis, as well as autophagy, was analyzed elaborately by Deng5 and his co-workers, in 2019. As most studies emphasized the apoptotic pathway, the least reports are found on autophagy. So, this study attempts to summarize the use of phytochemicals in various autophagy pathways treating oral cancer.
Autophagy in carcinoma pathway
Autophagy conserves as a lively interconnection in cell protection and a cytostatic association in carcinoma cell development6. The procedure introduced by the production of phagophore assemblage sites(PAS)6, Phosphatidylinositol 3-phosphate(PI3K) along with the endoplasmic reticulum/ER, was found to have a crucial function in the configuration of PAS7. Adenosine Monophosphate activated protein kinase(AMPK), mammalian target of rapamycin shortly termed ‘mTOR’ and unc-51 autophagy activating kinase-1(ULK1) found making easy phagophore development through autophagy initiation8, by the help of Vps15/p150, Vps34 and Beclin-1 in phagophore configuration9. The formation of phagophores, results in phagocytosis, consequently ending in elongation and sealing the membrane meant for the formation of autophagosome10. Adult autophagosomes attach with lysosomes, resulting in the development of autolysosomes2. Thus autolysosomes can be demolished by acidic hydrolases, help in additional recycling metabolism and consequently conserve cellular equilibrium.
However, mTOR is very significant in autophagy by defending or activating oncogenic cells. Chemotherapy drugs hold back cancer cells by altering the pathway of autophagy. So autophagy can be termed either a cellular existence or demise system11 and displays a vital position in maintaining metabolic adjustment in cancerous cells12. AMPK and mTOR have been recognized to be the most important signaling molecules that enhance autophagy through amino acids and the level of glucose8. Anyways, specified metabolites like palmitate, oxygen concentration, ATP to ADP ratio, particular levels of certain amino acids, ROS, growth factors as well as oncogenes control autophagy instigation and autophagosome construction. It has been confirmed by Youn13, that genes like Phosphatase and Tensin homolog deleted on chromosome 10(PTEN), Beclin-1, and Death associated protein kinase 1(DAPK 1) are cancer-suppressing and regulate the autophagy pathway. The expressed PTEN may endorse downregulation of the PI3K/AKT pathway, disturbing cancer augmentation by invigorating autophagy.
Development of autophagy in oral carcinoma
Autophagy is an actively impartial cellular process where unwanted nonfunctional cellular molecules break down or disintegrate by synthesis with lysosomes6; This cellular procedure plays a key role in regulating cell function and homeostasis. So autophagy conserves a dynamic relation in cell protection mechanism and a cytostatic linkage in tumor cell development6.
Autophagy-related genes like ATG7 take part in the covalent bonding between ATG5 – ATG12 in the membrane of autophagosome as an E1-like ubiquitination activase. Except, ATG8, recognized as microtubule-associated protein 1 light chain 3 (MAP1LC3 or LC3), is decisive for autophagy. Autophagy having dual activity can either enhance or restrict the onset of tumorigenesis. According to Saha14, autophagy may enhance the growth of tumor cells supplying nutrients in the later stage, still in the early stage, a long non-coding RNA FLJ22447 restricts autophagy of the cancer-associated fibroblasts (CAFs) and regulates the autophagic filth of IL33, where CAFs produce sufficient IL33 for the propagation of OSCC cells. Researchers like Zhang15 established that the neutrophil gelatinase-associated calcitonin (NGAL) gene activates mTOR, by blocking autophagy and enhancing OSCC. The mTOR protein kinase is a key downbeat controller of autophagy16 and controls numerous cell signaling pathways affecting cell growth, most of which with tyrosine kinase activity exhibit downstream growth factors. Structural activation of RAS, PI3K, AKT(activation mutation) and PTEN(inactivation mutations) are regularly found in cancer development. So Poillet-Perez and White17 suggested autophagy inhibition may endorse tumor intensification.
Then autophagy-mediated Oral carcinoma can be distinguished as ROS-dependent NUPR1-mediated autophagy, microRNA-mediated autophagy, or long Non-coding RNA-mediated autophagy. Autophagy in oral carcinoma treatment was established by various researchers in different ways. Some established CerS6 to enhance cisplatin-associated chemotherapy18, some regulated the ATG gene to block the oral carcinoma development and some tried to modify autophagy-related noncoding RNA to prevent oral carcinoma19.
Phytochemicals involved in cancer through autophagy
The Table. 1 gives a brief idea about some popular plant derivatives used in cancer therapy through autophagy.
Table 1: Some popular plant derivatives used in cancer therapy through autophagy
|
Sl. No. |
Name of the phyto chemicals |
Plant source |
Cancer type |
Signaling pathway |
References |
|
1 |
Gintonin (Glycoprotein) |
Panax ginseng |
Central Nervous System |
Akt/mTOR/p70S6K-mediated pathway |
20 |
|
2 |
Allicin( sulphur compound) |
Allium sativum |
Lung cancer |
A549 cells by ROS accumulation and facilitating S/G2-M phase arrest PI3K/mTOR signaling pathway |
21
22
|
|
3 |
Curcumin(polyphenolic compound) |
Curcuma longa L. |
Multiple cancers |
Autophagy in NSLCA549 cells Increased ROS and DNA damage, phosphorylation of ERK1/2 and p38 AMPK, inhibited Akt and P54 JNK |
23
24 |
|
4 |
Apigenin(Flavonoid) |
Justicia gendarussa |
Hepatocellular carcinoma |
PI3K/Akt/mTOR pathway
Kinase pathway/cell cycle at G2/M phase |
25
26 |
|
5 |
Aspalathin(polyphenolic compound |
Aspalathus linearis |
Prostate cancer |
AMPK and Fox pathways |
27 |
|
6 |
Hispolon (Polyphenol) |
Phellinus igniarius (L.) |
Naso-pharyngeal cancer |
ERK pathway |
28 |
|
7 |
Toxicarioside O |
Antiratoxicaria |
Colorectal cancer |
Akt/ |
29 |
|
8 |
Berberine(Alkaloid) |
turmeric, Oregon grape, goldenseal, and European barberry. |
Colon,Pancreas, Ovarian And breast cancer |
AMPK/mTOR/ULK1 pathway |
30 |
|
9 |
Celastrol(tri terpenoid) |
Tripterygium wilfordii |
Prostate cancer |
AR signaling pathway |
31 |
|
10 |
Evodiamine(quinolone alkaloid ) |
Evodia rutaecarpa |
Multiple cancer |
Beclin-1 and Bax expression for upregulation and Bcl-2 downregulation |
32 |
|
11 |
Fisetin(flavonoid) |
Strawberries, apples, persimmons, onions and cucumber |
Prostate cancer |
TOR signaling pathway |
33 |
|
12 |
Genistein (Isoflavon) |
legumes, |
Ovarian cancer |
Akt |
34 |
Phytochemicals affecting oral cancer therapy through autophagy
F.S. Yu35 reported on the cytotoxicity of tetrandrine over HSC-3 human oral cancer cells through autophagy and apoptosis. Tetrandrine improved LC3-I and -II expression initiating autophagy in HSC-3 cell lines. So tetrandrine-mediated autophagy in HSC-3 cells leads to cell fatality through PARP, caspases(3,8,9)/Becline I/LC3-I/II signaling pathways. The autophagy induced by tetrandrine through the Wnt/β-catenin pathway was also established by Zhang36. Chang37 for the first time revealed the resveratrol-mediated cell autophagy as well as apoptosis in cisplatin-resistant human oral tumor cells. They suggested resveratrol initiated autophagy vesicle formation, AVOs, and LC3B(Auto phagosome formation) in CAR cells. This also affects mRNA expression of genes like Beclin-1, LC3-II, Atg5, and Atg12, and is responsible for autophagy in CAR cells. This improves autophagy-involved proteins like Beclin-1, LC3-II, PI3K class III, 3-MA (an inhibitor of PI3K class III), and Atg complexes repressed the autophagic vesicle configuration by resveratrol. Chu38 demonstrated the action of thymoquinone(TQ) against 4 types of oral carcinoma cell lines (SAS, SCC-4, OC2, and SASVO3) of which SASVO3 cells were found to be mostly affected. Expression of autophagy-related proteins like Beclin-1, Rubicon, Class III, PI3K family, and Atg complex proteins, initiated autophagy by the formation of the autophagosome. Then LC3-I is attached to the lipid phosphatidyl ethanolamine and LC3-II is produced which is, an indicator of autophagy. mTOR is concerned with TQ-induced autophagy. Galangin stimulated autophagy by overexpressed genes like LC3I, LC3II, and Beclin 1 was demonstrated by Wang39 , which ended that galangin may cause human laryngeal carcinoma cell death, contributing to tumor suppression. Autophagy initiation was identified by Beclin-1 enhancement and p62 degradation which concluded that baicalein treatment induced autophagy in the OSCC cells. According to Li40, baicalein outstandingly amplified caspase-3 activity after repressing autophagic flux. This disclosed that baicalein-initiated autophagy reticence enhanced Cal27 cells to baicalein-initiated cell fatality by apoptosis. The autophagic pathways of the above-mentioned phytoconstituents are represented in Figure 1.
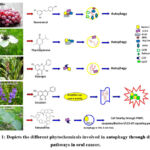 |
Figure 1: Depicts the different phytochemicals involved in autophagy through different pathways in oral cancer. |
Conclusions and Future Perspectives
Autophagy is an extremely multifaceted metabolic procedure that performs a decisive position in the body’s resistance to diseases. It shows a bifurcated effect over oral cancer. The outcome of autophagy on the incidence and expansion of oral cancer is mostly by the expression of autophagy-related genes. Plants and their bioactive products, which are rewards from mother nature to the human race, showed considerable anticancer activity and possess the capability to hold back the initiation and expansion of oral cancer, adopting mostly the apoptotic pathway. Phytochemicals in the pathway of apoptosis are widely studied in cancer therapeutics especially oral carcinoma, whereas the study of autophagy is almost neglected. That’s why there is a need for exploration of autophagic mechanisms and an explanation of the connection between the signaling pathway of autophagy and oral cancer. So we tried to put an insight into the process of autophagy in OSCC by the phytoconstituents which may help the researchers in the development of novel drugs.
Acknowledgment
We would like to thank IMS and SUM Hospital of SOA Deemed to be University for the use of their facilities. The authors are grateful to Professor Manojranjan Nayak, the president of SOA Deemed to be University, for supporting the study.
Conflict of Interests
The authors declare no conflict of interest.
Funding Sources
There is no funding Sources
Data availability
Data was collected from Scopus, Science direct, Elsevier, PubMed and Google Scholar.
Author contributions
SS performed literature searches and wrote the manuscript. SKB reviewed the manuscript. RB edited and designed the manuscript. All authors contributed to manuscript revision, and approved the submitted version.
Ethical approval
Not applicable.
References
- Santana-Codina N, Mancias JD, Kimmelman AC. The Role of Autophagy in Cancer. Annul Review Cancer Biology. 2017; 1:19-39. doi: 10.1146/annurev-cancerbio-041816-122338. PMID: 31119201; PMCID: PMC6527373
CrossRef - Kardideh B, Samimi Z, Norooznezhad F, Kiani S, Mansouri K. Autophagy, cancer and angiogenesis: where is the link?. Cell & bioscience. 2019; 9:1-0.
CrossRef - Wang X., Zhang H., and Chen X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resistance. 2019; 141–160. doi:10.20517/cdr.2019.10
CrossRef - Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Molecular pharmacology. 2014; 85(6):830-8.
CrossRef - Deng S, Shanmugam MK, Kumar AP, Yap CT, Sethi G, Bishayee A. Targeting autophagy using natural compounds for cancer prevention and therapy. Cancer. 2019; 125(8):1228-46.
CrossRef - Rahman MA, Rhim H. Therapeutic implication of autophagy in neurodegenerative diseases. BMB reports. 2017; 50(7):345.
CrossRef - Kotani T, Kirisako H, Koizumi M, Ohsumi Y, Nakatogawa H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proceedings of the National Academy of Sciences. 2018; 115(41):10363-8.
CrossRef - Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Molecular and cellular biology. 2012; 32(1):2-11.
CrossRef - Corona Velazquez AF, Jackson WT. So many roads: the multifaceted regulation of autophagy induction. Molecular and cellular biology., 2018; 38(21):e00303-18.
CrossRef - Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Current Biology. 2012; 22(1):R29-34.
CrossRef - Jung EM, Lim JH, Lee TJ, Park JW, Choi KS, Kwon TK. Curcumin sensitizes tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis through reactive oxygen species-mediated upregulation of death receptor 5 (DR5). Carcinogenesis. 2005; 26(11):1905-13.
CrossRef - Goldsmith J, Levine B, Debnath J. Autophagy and cancer metabolism. Methods in enzymology. 2014; 542:25-57.
CrossRef - Youn M, Gomez JO, Mark K, Sakamoto KM. RSK isoforms in acute myeloid leukemia. Biomedicines., 2021; 24;9(7):726.
CrossRef - Saha S, Zhang Y, Wilson B, Abounader R, Dutta A. The tumor-suppressive long noncoding RNA DRAIC inhibits protein translation and induces autophagy by activating AMPK. Journal of Cell Science. 2021; 134(24):jcs259306.
CrossRef - Zhang W, Yang S, Cui L, Zhang J. Neutrophil gelatinase-associated lipocalin worsens ischemia/reperfusion damage of kidney cells by autophagy. Renal Failure. 2016; 38(7):1136-40.
CrossRef - Viana SD, Reis F, Alves R. Therapeutic use of mTOR inhibitors in renal diseases: advances, drawbacks, and challenges. Oxidative medicine and cellular longevity. 2018; Oct 29.
CrossRef - Poillet-Perez L, White E. Role of tumor and host autophagy in cancer metabolism. Genes & development. 2019; 33(11-12):610-9.
CrossRef - Krasikova Y, Rechkunova N, Lavrik O. Nucleotide excision repair: From molecular defects to neurological abnormalities. International journal of molecular sciences. 2021; 22(12):6220.
CrossRef - Harini K. S, Ezhilarasan D. Promising autophagy inhibitors: Therapeutic implications in oral cancer. Oral oncology. 2022; 131:105948.
CrossRef - Rahman MA, Hwang H, Nah SY, Rhim H. Gintonin stimulates autophagic flux in primary cortical astrocytes. Journal of Ginseng Research. 2020; 44(1):67-78.
CrossRef - 21 . Pandey N, Tyagi G, Kaur P, Pradhan S, Rajam M. V, Srivastava T. Allicin overcomes hypoxia-mediated cisplatin resistance in lung cancer cells through ROS mediated cell death pathway and by suppressing hypoxia-inducible factors. Cell Physiology and Biochemistry. 2020; 54(4):748-66.
CrossRef - Sak K. Chemotherapy and dietary phytochemical agents. Chemotherapy research and practice. 2012; 2012.
CrossRef - Liu F, Gao S, Yang Y, Zhao X, Fan Y, Ma W, Yang D, Yang A, Yu Y. Antitumor activity of curcumin by modulation of apoptosis and autophagy in human lung cancer A549 cells through inhibiting PI3K/Akt/mTOR pathway. Oncology reports. 2018; 39(3):1523-31.
CrossRef - Masuelli L, Benvenuto M, Di Stefano E, Mattera R, Fantini M, De Feudis G, De Smaele E, Tresoldi I, Giganti MG, Modesti A, Bei R. Curcumin blocks autophagy and activates apoptosis of malignant mesothelioma cell lines and increases the survival of mice intraperitoneally transplanted with a malignant mesothelioma cell line. Oncotarget. 2017; 8(21):34405.
CrossRef - Yang J, Pi C, Wang G. Inhibition of PI3K/Akt/mTOR pathway by apigenin induces apoptosis and autophagy in hepatocellular carcinoma cells. Biomedicine & Pharmacotherapy. 2018; 103:699-707.
CrossRef - Zhong Y, Krisanapun C, Lee SH, Nualsanit T, Sams C, Peungvicha P, Baek SJ. Molecular targets of apigenin in colorectal cancer cells: involvement of p21, NAG-1 and p53. European Journal of Cancer. 2010; 46(18):3365-74.
CrossRef - Johnson R, Shabalala S, Louw J, Kappo AP, Muller CJ. Aspalathin reverts doxorubicin-induced cardiotoxicity through increased autophagy and decreased expression of p53/mTOR/p62 signaling. Molecules. 2017; 22(10):1589.
CrossRef - Hsin MC, Hsieh YH, Wang PH, Ko JL, Hsin IL, Yang SF. Hispolon suppresses metastasis via autophagic degradation of cathepsin S in cervical cancer cells. Cell death & disease. 2017; 8(10):e3089-.
CrossRef - Huang YH, Sun Y, Huang FY, Li YN, Wang CC, Mei WL, Dai HF, Tan GH, Huang C. Toxicarioside O induces protective autophagy in a sirtuin-1-dependent manner in colorectal cancer cells. Oncotarget. 2017; 8(32):52783.
CrossRef - Wang J, Qi Q, Feng Z, Zhang X, Huang B, Chen A, Prestegarden L, Li X, Wang J. Berberine induces autophagy in glioblastoma by targeting the AMPK/mTOR/ULK1-pathway. Oncotarget. 2016; 7(41):66944.
CrossRef - Guo J, Huang X, Wang H, Yang H. Celastrol induces autophagy by targeting AR/miR-101 in prostate cancer cells. PLoS One. 2015; 10(10): e0140745.
CrossRef - Rasul A, Yu B, Zhong L, Khan M, Yang H, Ma T. Cytotoxic effect of evodiamine in SGC-7901 human gastric adenocarcinoma cells via simultaneous induction of apoptosis and autophagy. Oncology reports. 2012; 27(5):1481-7.
- Suh Y, Afaq F, Khan N, Johnson J. J, Khusro F. H, Mukhtar H. Fisetin induces autophagic cell death through suppression of mTOR signaling pathway in prostate cancer cells. Carcinogenesis. 2010; 31(8):1424-33.
CrossRef - Gossner G, Choi M, Tan L, Fogoros S, Griffith KA, Kuenker M, Liu JR. Genistein-induced apoptosis and autophagocytosis in ovarian cancer cells. Gynecologic oncology. 2007; 105(1):23-30.
CrossRef - Yu FS, Yu CS, Chen JC, Yang JL, Lu HF, Chang SJ, Lin MW, Chung JG. Tetrandrine induces apoptosis via caspase‐8,‐9, and‐3 and poly (ADP ribose) polymerase-dependent pathways and autophagy through beclin‐1/LC3‐I, II signaling pathways in human oral cancer HSC‐3 cells. Environmental toxicology. 2016; 31(4):395-406.
CrossRef - Zhang Z, Liu T, Yu M, Li K, Li W. The plant alkaloid tetrandrine inhibits metastasis via autophagy-dependent Wnt/β-catenin and metastatic tumor antigen 1 signaling in human liver cancer cells. Journal of experimental & clinical cancer research. 2018; 37(1):1-1.
CrossRef - Chang CH, Lee CY, Lu CC, Tsai FJ, Hsu Y. M, Tsao JW, Juan YN, Chiu HY, Yang JS, Wang CC. Resveratrol-induced autophagy and apoptosis in cisplatin-resistant human oral cancer CAR cells: A key role of AMPK and Akt/mTOR signaling. International journal of oncology. 2017; 50(3):873-82.
CrossRef - Chu SC, Hsieh YS, Yu CC, Lai YY, Chen PN. Thymoquinone induces cell death in human squamous carcinoma cells via caspase activation-dependent apoptosis and LC3-II activation-dependent autophagy. PloS one. 2014; 9(7): e101579.
CrossRef - Wang HX, Tang C. Galangin suppresses human laryngeal carcinoma via modulation of caspase-3 and AKT signaling pathways. Oncology Reports. 2017; 38(2):703-14.
CrossRef - Li B, Lu M, Jiang XX, Pan MX, Mao JW, Chen M. Inhibiting reactive oxygen species-dependent autophagy enhanced baicalein-induced apoptosis in oral squamous cell carcinoma. Erratum: January 2021; 75 (1), p. 259.
CrossRef








