Sarab Mohammed Mahdi1 , Mais Emad. Ahmed2
, Mais Emad. Ahmed2 and Adawia Fadhil Abbas1
and Adawia Fadhil Abbas1
1Department of Biology/College of Education Pure Science/University of Diyala, Iraq
2Department of Biology/College of Science/University of Baghdad, Iraq
Corresponding Author E-mail: mais.emad@sc.uobaghdad.edu.iq
DOI : https://dx.doi.org/10.13005/bpj/2939
Abstract
Urinary tract infections linked to catheters are believed to be caused most frequently by Proteus mirabilis. It produces urease, which greatly increases the potency of catheter occlusion caused by swarming. Pathogenic bacteria use swarming as one of their main virulence mechanisms to evade antibiotics; as a result, there is an increasing need to develop novel antibiotic substitutes. Investigating the possible antibiofilm capabilities of artificial zinc oxide nanoparticles (ZnO NPs) made from E. Faecium was the aim of this study. By generating reductive enzymes, bacterial cells are able to catalyze the biosynthesis process. Bacteriocin-like inhibitory substance (BLIS) was used to create the nanoparticles. AFM, TEM, FESEM, and other analytical tools were used to characterize the synthesized zinc nanoparticles and determine the chemical and physical characteristics of the products. Weak swarming is shown by microorganisms that develop strong swarming. After incubation, the ZnO nanoparticles were incubated for 24 or 48 hours at 37°C at a sub-MIC of 32 µg/ml. After these isolates were treated with zinc nanoparticles, downregulation of rsbA expression was detected via real-time PCR compared to that in the untreated isolates. Zinc oxide nanoparticles can serve as antibacterial agents in a concentration-dependent manner, according to all of the study's findings. This was demonstrated by the notable downregulation of rsbA gene expression, which effectively inhibits the production of biofilms and swarming motility. This was demonstrated by their noteworthy downregulation of rsbA gene expression, which effectively promoted swarmed motility.
Keywords
BLIS; P. mirabilis; rpoA; rsbA; ZnONPs
Download this article as:| Copy the following to cite this article: Mahdi S. M, Ahmed M. E, Abbas A. F, Effect of Enterococin – Zinc Oxide Nanoparticles on Gene Expression of rsbA Swarming Genes in Proteus mirabilis isolation Catheter Urine. Biomed Pharmacol J 2024;17(2). |
| Copy the following to cite this URL: Mahdi S. M, Ahmed M. E, Abbas A. F, Effect of Enterococin – Zinc Oxide Nanoparticles on Gene Expression of rsbA Swarming Genes in Proteus mirabilis isolation Catheter Urine. Biomed Pharmacol J 2024;17(2). Available from: https://bit.ly/44Rv7Ec |
Introduction
Proteus mirabilis is rod-shaped motile bacterium that belongs to gram-negative and facultative anaerobic species. P. mirabilis is a common causative agent of urinary tract infection (UTI) in the intricate urinary tract, most commonly in patients with Indwelling catheters , The organism shows swarming motility and urease activity.1 P. mirabilis grows on an agar surface for a riodof time (which varies depending on the medium,humidity and temperature), at which point it differentiates into swarm cells and proliferates as a population. Hauser originally described P. mirabilis’s capacity to swarm across solid surfaces in 1885 as an organized group P. mirabilis develops into very long, multinucleate, highly motile hyperflagellated cells during the swarm process.2 During the consolidation phase, swarm cells periodically slow down or cease to move and dedifferentiate into shorter rod-shaped cells. The bull’s-eye pattern is the result of repeated rounds of swarming and consolidation.3 Flagellar function is the primary determinant of swarming motility. There is a hierarchy in the expression of the genes involved in flagellar biosynthesis,with a master regulatory transcription factor (or “master regulator”) controlling the expression at the top. Master regulators control the synthesis of flagellar basal bodies, stimulate the expression of flagellar genes, and act as an integrating point for environmental signals. The formation of flagellar membranes is intricate, and there exist species-specific transcriptional and posttranscriptional regulatory systems.4 Swarming behavior Is partially controlled by Rsba gene product. rsbA may operate as a protein sensor of environmental circumstances, and rsbA was stimulated swarming production.5 The swarming regulation gene, rsbA . Due to its distinct features and the noteworthy importance of nanoparticles, it has emerged as the most innovative, cutting-edge, and well-known area of study in contemporary science. Materials science and healthcare both make substantial use of nanoparticles. Their innovative solutions in various scientific domains have led to their unexpected rise in prominence in recent years.6 With respect to their macroscale counterparts, they are distinct from bulk materials due to their unique physicochemical features (such as color, dispersion, and thermodynamics) and high surface area-to-volume ratio.7
Nicin A , such as Bacteriocin Produced from lactic acid bacteria, it provides additional protection and a crucial component for research and development of sustainable biotechnologies,including food preservatives for human health and environmental preservation.8 According to.9 Onother hand foucus synergestic bacteriocin-nanoparticales syntersis from different method have shown to be a workable solution and the most potent antibacterial agent in vitro and invivo.10 According to11 the biogentic fro synthesis silver nanoparticales from peel lemone green synthesizes and friendly to evnvironent Zinc oxide nanoparticles, or ZnO-NPs, are an important and versatile inorganic molecule that belong to the class of metal oxide nanomaterials due to their unique physical and chemical characteristics. They exhibit good photostability,a high electrochemical coupling coefficient, an extended radiation absorption spectrum, and high chemical stability. Their molecular formula is ZnO.12 Because of their small size, ZnO particles at the nanoscale exhibit potent antibacterial effects. These particles can initiate a number of bactericidal activities, including those in the bacterial surface or bacterial core,once they are within the bacterial cell.13 These findings align with other studies that looked at how certain nanoparticles, such zinc oxide, affected the movement of swarming gram-negative bacteria like P. mirabilis. As stated by.14
Materials and Methods
Sample collection
Proteus mirabilis
Between September 2023 and November 2023, patients were referred to the Al-Yarmouk Teaching Hospitals in Baghdad, where urine samples were taken from various sources. There were 150 patients with symptoms of a urinary tract infection. Catheter urine collected midstream was transferred straight to the laboratory for culture in sterile containers. Samples were grown on blood agar and MacConkey agar, a selective and differentiation medium used in conjunction with the previously mentioned media, to establish preliminary identification. The cultures were incubated at temperature 37°C and for twenty-four hours 15.
Isolation and identification:
Initial diagnoses depended on the phenotypic characteristics of a colony, including its shape, swarming phenomenon, size, color, texture, and arrangement. All P. mirabilis isolates were numbered from (M1 to M70). Proteus spp. appear as pale colonies on MacConkey agar, and swarming phenomena appear on blood agar plates after overnight incubation at temperature 37°C16. The results were confirmed by the VITEC 2 system.
Biochemical tests
Several standard biochemical tests were performed to diagnose bacterial isolates according to17
Indole test
Bacterial colonies were inoculated into test tubes containing peptone water medium, incubated for 24 hours at temperature 37°C, and then 1-2 drops of Kovacs reagent were gently shaken into the medium. The appearance of a red ring (indole ring) on the medium’s surface indicates a positive test.
Urease Production test
This test was used to detect bacteria’s ability to produce urease, an enzyme that converts urea to ammonia and carbon dioxide. The Stab method was used to streak bacterial isolates onto slanted urea agar medium, which was then incubated at temperature 37°C for 24 hours. As the medium changes color, pink indicates a positive test result and yellow indicates a negative test result.
Motility test
The movement medium was inoculated with bacterial isolates in the shape of a stab , and the tubes were incubated for 24 hours at temperature 37°C. Bacterial movement is evidenced by the spread of growth around the stab site.
Detection of Swarming motility
All P. mirabilis isolates were injected into 5% sheep blood agar plates (SBAs), and after 24 hours at temperature 37°C to allow for revival, they were cultured. The next day, the turbidity of a single colony growing on SBA was reduced to 108 CFU/ml, or 0.5 McFarland standard, by diluting it in a saline solution. A saline bacterial solution containing 107 CFU was produced in 100 μl and used to inoculate several SBA plates. P. mirabilis swarming motility was observed by inoculating droplets of the bacterial suspension in the center of the plate. The next day, each isolate’s swarming motility was visually evaluated.
Detection of the rsbA gene
The tested gene was amplified using a primer and traditional PCR. The sequence was obtained fromthe references listed in the table(1) 1819. PCR amplification was carried out in 20 µl volumes containing 10 µl of GoTaq Green Master Mix (2X), 1 µl of primer (10 pmol), 6 µl of nuclease-free water, and 2 µl of template DNA. PCR Express (Thermal Cycler, Thermo Fisher Scientific, USA) was used for PCR cycling. The following temperature schedule was used: 5 minutes of initial denaturation at 95°C, followed by 1 cycle of denaturation at 95°C for 30 seconds, annealing at 50, 55, or 60°C for 30 seconds, and extension at 72°C for 30 seconds. The last extension step was run for seven minutes at 72°C, after which the reactions were stopped by a ten-minute incubation at 10°C. The rsbA primer used is indicated in Table (1).
Table 1: Primers used in this research.
|
Primer Name |
Sequence 5’-3’ |
Annealing Temp.(°C) |
References |
|
rsbA-F |
CTATACCTACCGCACCATGT |
|
18 |
|
rsbA-R |
GAAGTCCCATCCGTTGATAC |
60 |
|
|
rpoA-F |
GCGTGTTATAGCCCAGTTGA |
|
19 |
|
rpoA-R |
AGGCTGACGAACATCACGTA |
|
|
Sample collection and bacterial identification of Enterococcus faecium
This study used 25 samples collected from patients visiting a private dental clinic from the gums. Samples were collected using transport media, then kept in a cool place until transported to the laboratory. Samples were then cultured in MRS broth. Then on MRS agar and incubated for 24 hours at temperature 37°C. The isolates were then numbered (E1–E25) and cultured on MRS agar. Several tests have been used to diagnose. . (20)
Screening for BLIS production by E. faecium
Bacteriocin-like inhibitory substance (BLIS)
In samples obtained from MRS broth culture, the antimicrobial spectrum synthesized by the isolated lactic acid bacteria was analysed. After 24 h of incubation at 37°C and under conditions of an appropriate pH (5.30), yeast extract (1%), glucose (1%) and peptone (1%), the BLIS were recovered by centrifugation a speed 6000 rpm for 15 min (21) and then transferred to new tubes for various tests (22). The isolate with the highest bacterial yield was selected using the filtration paper disk (FPD) method.
Filter Paper Disc Method (FPD)
To test for the presence of inhibitory substances, bacteria were spread on the surface of MHA plates with the BLIS preparation. Five millimeter diameter sterile filter paper discs saturated with 100 µl of BLIS suspension were placed on MHA plates(23). The plates were subsequently incubated aerobically at temperature 37°C for 24 to 48 hours after leaving the plates two hours at room temperature. The inhibition zones that formed around the paper discs were measured and recorded.
Solutions used for protein determination (Bradford, 1976)24
The protein reagent Coomassie Brilliant Blue G-250 (100 mg) was reconstituted in 50 millilitres of 95% ethanol. Phosphoric acid (85% (w/V) in 100 ml) was added to this solution. A final volume of one liter was achieved by diluting the resultant solution. Brilliant Blue G-250, 4.7% (w/v) ethanol, and 8.5% (w/v) phosphoric acid were used.
Partial purification of the bacteriocin-like inhibitory substance
Partially purified crud bacteriocin was prepared via precipitation with ammonium sulfate at different concentrations 25.
Ammonium sulfate precipitation
Precipitation of ammonium sulfate was achieved by gradually adding (5 g) ammonium sulfate to the crude enzyme with continuous stirring on ice at different degrees of saturation. After that, the mixture was centrifuged for 20 minutes at 4°C at 6,000 rpm. When the precipitate at each concentration was dissolved in the appropriate volumes of phosphate buffer solution, the supernatant was discarded. The optimal saturation ratio was ascertained by measuring the enzyme activity. 26
A stock solution of Zinc acetate [Zn.2H2O(CH3CO2)2] was prepared by dissolving 0.01 gm of Zn.2H2O(CH3CO2)2 in 50 ml deionized water 27.
ZnoNP Biosynthesis
For the synthesis of the Zn NPs, 7 ml of BLIS bacteria was added to 3 ml of 1 mM Zn.2H2O(CH3CO2)2 (final concentration) at room temperature with a pH of 5. The flasks were incubated at 30°C to 35°C for three days, and each color change was recorded 28. After incubation, the reaction mixture used to prepare the ZnO NPs was centrifuged after 27 hours of incubation, and the precipitate was collected and washed three times with deionized water. The collected nanoparticles in the form of pellets were transferred to a hot air oven set at 120°C to evaporate all the liquid. The powdered dehydrate was meticulously collected and preserved for additional examination. 29 The formation of nanoparticles can be identified by the change in color. 30
Characterization of ZnO NPs: This study was performed in the Department of Chemistry/College of Sciences laboratories at the University of Al-Nahrain.
UV‒Visible (UV‒VIS) spectral analysis
The formation of nanoparticles was confirmed by measuring the wavelength of the reaction mixture in a 2 ml quartz cuvette with a path length of 1 cm in the UV‒VIS spectrum of a PerkinElmer spectrophotometer with a resolution of Inm. The samples were scanned from 200 -800 nm at a speed of 500 nm/min with a blank reference used for spectrophotometer correction31.
Atomic force microscopy (AFM) analysis
We used atomic force microscopy to measure the average diameter of the generated nanoparticles. A few drops of the manufactured NPs were applied to a silica glass plate and allowed to dry at room temperature in the dark to produce a thin layer of the material. AFM was then used to scan the deposited glass film plate. 32
X-ray diffraction (XRD)
X-ray diffraction (XRD) was used to determine the crystal structure of the NPs. There are several methods for grinding powdered samples via XRD, depending on the sample matrix, sample size, and/or amount of generated material needed. 33
Energy dispersive X-ray (EDX) spectroscopy
Energy dispersive X-ray spectroscopy (EDX) can be used to determine the elemental makeup of materials, including nanoparticles. When a sample is exposed to a high-energy X-ray, EDX reveals the distinctive X-rays that the elements inside the sample release into the air34.
Field emission scanning electron microscopy (FESEM)
Field scanning electron microscopy (FESEM) was used to examine the morphology of the synthesized nanoparticles. To generate images, an electron beam was used to scan the sample surface. The electrons in the sample interact with the atoms and surface topography.35
Antibacterial activity of ZnONPs
The microdilution method was used to determine the minimum inhibitory concentrations (MICs) of ZnONPs against P. mmirabilis, which was cultured in the appropriate medium for a full night. We dissolved and diluted the ZnONPs. Different concentrations of ZnONPs (1000, 500, 250, 125, and 62.5 μg/mL) were prepared. P. mmirabilis on Mueller–Hinton agar was cultured, and an agar well diffusion assay was used to test the presence of ZnONPs36
The minimum inhibitory concentration (MIC) of the ZNO NPS was determined.
The microdilution method can be used to determine the minimum inhibitory concentration (MIC) in culture broth. Coagulants varying in concentration from 1000, 500, 250, 125, and 42.5 g/ml were produced. One hundred litres of Mueller-Hinton broth was added to the first column of a 96-well microtiter plate, and an additional 100 litres were added to each of the remaining wells. Next, 100 litres of each dilution were added to each well, bringing the total capacity to 200 litres. Each well received ten litres of a modified P. mirabilis37 bacterial isolate. Microtiter plates were covered and incubated at 37°C for an entire day.
RT‒qPCR protocol.
RNA purification
Following the TRIzolTM Reagent procedure, RNA was extracted from the sample using the following steps:
Sample lysis
Suspension-grown cells: After centrifuging the culture for two minutes at 13,000 rpm, the supernatant was removed, and 0.5 mL of TRIzolTM Reagent was added to the pellet. The lysate was homogenized by pipetting repeatedly.
For three-phase separations
The lysate was mixed with 0.2 mL of chloroform in each tube, and the tube cap was then fastened.
The mixtures were separated into a lower organic phase, interphase, and a colorless upper aqueous phase after being incubated for two to three minutes and centrifuged for ten minutes at 12,000 rpm.
A fresh tube was filled with the RNA-containing aqueous phase.
For RNA precipitation
The aqueous phase was mixed with 5 mL of isopropanol, incubated for 10 minutes, and then centrifuged for 10 minutes at 12,000 rpm.
The supernatant was discarded when total RNA precipitated, and a white gel-like pellet formed at the bottom of the tube.
For RNA washing
For each tube, 0.5mL of 70% ethanol was added and vortex briefly then centrifuged for 5 minutes at 10000 rpm.
Ethanol then aspirated and air-dried the pellet.
For RNA solubility
Pellet was rehydrated in 50 µl of Nuclease Free Water then incubated in a water bath or heat block set at 55-60°C for 10-15 minutes.
Determination of RNA and cDNA yields
The Quantus Fluorometer was used to evaluate sample quality for downstream applications by measuring the concentration of extracted RNA or cDNA. 199 µl of diluted QuantiFlour Dye was mixed with 1 µl of RNA or CDNA. RNA concentrations were measured after a 5 min incubation period at room temperature in a dark place.
Table 2: Synthesis of cDNA from RNA using primers for rsbA and rpo
|
Master mix components |
Volume |
|
qPCR Master Mix |
5 ul |
|
RT mix |
0.25 ul |
|
MgCl2 |
0.25 ul |
|
Forward primer |
0.5 ul |
|
Reverse primer |
0.5 ul |
|
Nuclease Free Water |
2.5 ul |
|
RNA |
1 ul |
|
Total volume |
10 ul |
Results
Identification of Bacterial Isolates
Proteus mirabilis isolates
A total of one hundred and fifty specimens were collected during the period between September and December 2023 from different urine sources, including urine samples from UTI patients (70) and catheter urine (80). All specimens were directly inoculated on MacConkey agar and blood agar plates. Of the total number of specimens, 70 (46%) were identified as P. mirabilis, as shown in Table (3). Furthermore, 30 (42.85%) cultured urine samples were diagnosed with P. mirabilis strains. Additionally, 40 (50%) of the catheter urine sample cultures were identified as P. mirabilis (Figure 1).
Table 3: Prevalence of Proteus mirabilis isolates among the samples
|
Source of |
No. of |
No. (%) |
No.of |
|
Catheter urine |
80 |
40(50%) |
10(12.5%) |
|
Urine |
70 |
30(42.85%) |
5(7.14%) |
|
Total |
150 |
70 (46%) |
15(10%) |
 |
Figure 1: Incidence of P. mirabilis in collected samples |
Biochemical tests for Proteus mirabilis.
Different biochemical tests were performed for each isolate, including the catalase test, urease test, motility test, which yielded positive results. The indole test yielded negative results.
Detection of Swarming motility
All Pmirabilis isolates were cultivated on blood agar to test their capacity for extravasation. After one day, 40 isolates (66.6%) exhibited narrative movement, according to the results. Figure (2) Reliance on genetics. The isolates (M8 and M3) were found to be the most potent in creating the swarm phenomenon. mirabilis can swarm on various catheter surfaces, and because of this, it has been observed to move from the associated urinary tract infection (CAUTI) through the periurethral skin, the catheter, and the bladder.
 |
Figure 2: Swarming motility of P. mirabilis on blood agar at 37°C for 24 h. incubation. |
Polymerase chain reaction (PCR)
PCR revealed that ten P. mirabilis isolates containing the rsbA gene were amplified; these isolates were chosen due to their multidrug resistance (MDR) status. After electrophoresis on a 1.5% agarose gel stained with ethidium bromide, at 75 volts for 50 minutes, and under an ultraviolet (UV) trans illuminator, the positive gene result was subsequently confirmed. The present study revealed the presence of a sharp, singular, and nondispersed 180 bp MexB gene band, which was clearly distinguished from the DNA ladder, as demonstrated in Figure (3). Notably, there was no evidence of DNA degradation, as indicated by the absence of any smearing of the gene band.
 |
Figure 3: Results of the amplification of rsbA genes of bacterial species were fractionated on 2% agarose gel electrophoresis stained with Eth.Br. |
Identification of Bacterial Isolates of E. faecium
Isolation of E. faecium In this study, twenty-five specimens of E. faecium from the gums were collected . were examined, and only 10 isolates were identified, as shown in Table 4.
Table 4: Prevalence of E. faecium isolates among the samples
|
Source of samples |
Other gram -negative bacteria No. Isolates |
No. of E. faecium |
|
Gums |
15 |
10 (66.6%) |
|
Total |
25 |
10(40%) |
Screening of the Antimicrobial Activity of E. faecium
The bacterial productivity of BLIS was investigated using the filter paper disc method (FPD) to discriminate the isolates that possessed the capacity to produce an inhibitory substance (BLIS). The primary screening revealed that approximately five isolates of E. faecium produced inhibitory substances, and inhibition zones between 10 mm and 26 mm were recorded. After secondary screening, the greatest inhibition diameter of the isolate was detected (E20). The 5 isolates were generally screened for BLIS production utilizing filter paper disk, and the resulting isolate was named E. faecium (E20) was the most productive isolate, and the best result was obtained with the filter paper disk method. The best productivity was observed at pH 5, and the optimal incubation conditions were 72 hours. The optimal nitrogen source was 1% yeast extract, and the optimal carbon source (glucose and peptone) was 1%.
Protein Determination
The protein concentrations in the four isolates were 12.81 mg/ml for E3, 12.43 mg/ml for E11, 12.32 mg/ml for E5, and 36 mg/ml for E20. The E20 isolate was the most productive.
Partial purification of the bacteriocin-like inhibitory substance
The bacteriocins generated by E. faecalis were partially purified using a step-gradient elution assay with ammonium sulfate in 100 mM phosphate buffer (pH 5). The experimental model must be followed when bacteriocins are applied as crude, partially purified preparations.
Zinc Oxide Nanoparticle Biosynthesis:
In the present study, zinc acetate solution and E. Faecuim metabolic filtrated extract were mixed for 20 minutes before a white precipitate became visible to the unaided eye, marking the first visual observation of the ZnO-NPs. The appearance of this white precipitate was caused by the presence of zinc hydroxide, which was coated with the metabolic filtrate of E. faecium. The second stage involved drying zinc hydroxide to create ZnO-NPs. Figure (4).
 |
Figure 4: Zinc nanoparticle biosynthesis A: After incubation, the cell-free extract was incubated with zinc acetate B: Nanoparticle sedimentation C: Drying of zinc oxide nanoparticles. |
Characterization of ZnONPs
UV‒Vis spectral analysis
UV‒visible spectroscopy was utilized to measure the optical characteristics of the ZnONPs in the 200-800 nm range. As the particle size decreases, the intensity of the absorption peak shifts (blueshifts) toward a lower wavelength. When ZnONPs are stimulated by UV light, their valence electrons show an absorption peak in the 267 nm spectrum, indicating a characteristic band for pure ZnO. Figure (5).
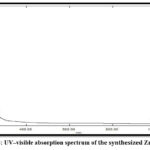 |
Figure 5: UV‒visible absorption spectrum of the synthesized ZnO NPs |
Atomic force microscopy (AFM) analysis
We can learn more about the topography and roughness of nanoparticles through AFM investigations. The AFM 3D image shown in Figure (6) shows the smooth surfaces of different types and sizes of nanoparticles. Round and triangular artificial nanoparticles are produced
 |
Figure 6: 2D AFM image of ZnO NPs biosynthesized by S. mutans |
Field emission scanning electron microscopy (FESEM)
SEM was used to analyse the morphological characteristics of the biosynthesized ZnO-NPs. Figure 11. Samples subjected to two separate magnifications during scanning. The as-prepared ZnO-NPs were found to have distinct architectures and rough surfaces based on the scanned samples. Moreover, the number of aggregated particles was greater. Overall, tiny ZnO-NPs were effectively created by the metabolic filtrated extract of E. faecium.
 |
Figure 7: SEM images of ZnO NPs |
Energy dispersive X-ray (EDX) spectroscopy
Energy dispersive X-ray analysis (EDX) was used for analysis. Fig. 8 shows the presence of zinc and oxygen in the ZnO-NPs. Moreover, other components, such as nitrogen, carbon, phosphorus, and sulfur, are predominantly attributed to the metabolic filtrated extract of E. faecium.
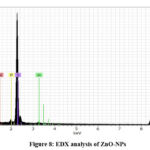 |
Figure 8: EDX analysis of ZnO-NPs |
X-ray diffraction (XRD):
XRD is a powerful technique that provides information about the structure, average size and crystalline nature of a sample. Figure (9) shows the XRD pattern of the ZnO NPs synthesized using E. faecium.
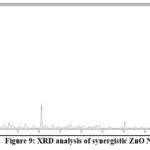 |
Figure 9: XRD analysis of synergistic ZnO NPs. |
Antibacterial activity of ZnONPs
According to the MIC results and the diameters of the zones of inhibition observed in Figure 14, the ZnO nanoparticles had significant antimicrobial activity against P. mmirabilis bacteria isolated from urine and catheter urine. Further evidence of a statistically significant difference (p < 0.05) in the disseminated mean zone of bacterial inhibition following treatment with varying concentrations of ZnONPs (1000, 500, 250, 125, and 62.5 μg/mL) is shown in Fig. 10. The statistical analysis revealed a significant difference (p < 0.05) in the mean zone of bacterial inhibition after treatment with 1000 µg/mL ZnNPs. There were significant differences between the P. mmirabilis isolations.
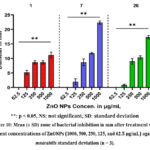 |
Figure 10: Mean (± SD) zone of bacterial inhibition in mm after treatment with different concentrations of ZnONPs (1000, 500, 250, 125, and 62.5 μg/mL) against P. mmirabilis standard deviation (n = 3). |
Minimum inhibitory concentration of ZnO NPs
The MICs of ZnO NPs were determined for P. mirabilis isolates (P.m2, P.m3, P.m4, P.m5, P.m6, P.m7 and P.m8). Serial dilutions of each nanoparticle (1000, 500, 250, 125 and 64 µg/ml) were introduced into the bacterial broth tube. The different MIC values were as follows: ZnO NPs had a significant effect on bacterial cell viability and resulted in 100% viability at 1000 µg/ml, while the MIC of ZnO NPs was 250 µg/ml for the seven isolates. The sub-MIC of the ZnO NPs was 125 µg/ml. The sub-MIC of each nanoparticle was used to determine the inhibitory effect on swarming motility
Effect of ZnO NPs on rsbA gene expression
Real-time PCR was used to determine changes in rsbA gene expression after exposure to ZnO NPs at sub-MICs (250 µg/ml). The expression of the RsbA gene was significantly downregulated in the M8 isolate, while the expression of the RsbA gene was upregulated in the M3 isolate. Several studies have confirmed the decreased folding of rsbA after exposure to ZnO NPs and other nanoparticles. studied the impact of zinc oxide and silver nanoparticles on the expression of the rsbA genes involved in the swarming phenomenon in P. mirabilis and discovered a decrease in gene expression for isolates after treatment compared to pretreatment gene expression. Figure 11 shows that the expression of the rsbA gene was significantly (p <0.01) downregulated after bacterial CuO-ZnO NP treatment compared with that in the control (no treatment). This study may be the first to investigate the possible impact of ZnO NPs on these genes, even though no research has investigated how ZnO NPs affect the expression of the rsbA gene in P. mirabilis. It should be noted, however, that other nanoparticles were used to target genes related to the regulation of quorum sensing in this species.
 |
Figure 11: Mean ± SD of the fold change in the gene expression of rsbA before and after treatment with ZnO NPs compared with that in untreated cells (n =3). |
Discussion
Using the VITEK 2 system, seventy isolates of P. mirabilis were discovered in addition to swarming on blood agar and forming colonies on MacConkey agar. One feature of P. mirabilis that is easily noticeable on firm agar surfaces is the bull’s-eye pattern. Proteus mirabilis, a human opportunistic pathogen, and other highly resistant bacteria concurred with these results(38). Various biochemical tests were conducted on P. mirabilis isolates and showed positive results for catalase, urease, and methyl red and motility tests and negative results for indole oxidase tests. These results are similar to (39). The results of this study are consistent with studies confirming that P. mirabilis possesses bull’s-eye swarming motility that enables bacteria to rapidly migrate through all types of catheters through their numerous flagella and easily spread the infection to other parts of the urinary system(40). The antibacterial activity of the BLIS suspension was studied using the disk diffusion method to distinguish isolates that possess the ability to produce the inhibitor. According to a different study, the filter paper disk test has numerous benefits over alternative techniques, including simplicity, affordability, the capacity to test a large number of bacteria and antimicrobial compounds, and ease of interpreting the results. (41). The generation of a light yellow to white precipitate served as an indicator of nanoparticle production. Following centrifugation, the precipitate turned white and was collected as a white powder after being microwave-dried. In recent years, bacteria have been used to create nanomaterials with exceptional properties, primarily gold, silver, and zinc nanoparticles, for the creation of antimicrobials with in vitro activity against harmful bacteria(42). UV spectroscopy was used to verify the biogenesis of the ZnO NPs. The generation of freshly generated ZnO NPs with a maximum peak at 267 nm was confirmed by the results. This is in line with research showing that ZnO NPs have a restricted distribution of sizes because they are nanoscale particles, which causes a sharp absorption peak to be visible. Because zinc oxide nanoparticles absorb light well in the 200–400 nm UV range, they are appropriate for use in medical applications(43). In this study, AFM analysis of ZnO NPs was performed using scanning microscopy (CSPM) to identify and characterize the nanoparticle distributions. The estimated grain size and average square roughness were determined. Microbial synthesis of ZnO nanoparticles can produce products with different sizes, shapes, activities and behaviors. These differences can be attributed to the synthesis pathway, enzymes used by the microorganisms, temperature, and other biological factors(44). Compared with chemical methods, the size of biosynthesized zinc oxide NPs can be larger(45). To determine the morphology, size, and elemental and structural content of the NP samples, FESEM analysis was carried out. FESEM images from a previous study revealed that they were essentially spherical and uniform in appearance with a diameter of 15 to 19 nanometers. Compared to EDX, FESEM allows the determination of the presence of different components in the examined model(46). The XRD patterns showed clear peaks that matched the diffraction peaks at 20 Hz, confirming the generation of ZnO NPs. The composite material was in the nanoscale range, as indicated by the broadening of the diffraction peak line (47). The MIC of ZnO NPs in this study was 250 µg/ml. The sub-MIC of ZnO NPs was 125 µg/ml. The sub-MIC of each nanoparticle was used to determine its inhibitory impact on motility during swarming. Numerous studies have documented the anti-biofilm activity of zinc oxide NPs; one study revealed that treating Pseudomonas aeruginosa bacteria with zinc oxide NPs significantly reduced the ability of bacteria to produce biofilms(48). Compared with other metal nanoparticles, nanoparticles exhibit enhanced properties that improve their activity, such as a larger surface area, smaller band gap, smaller particle size, and greater stability, according to recent research on this topic(49). Real-time PCR technology was used to determine changes in rsbA gene expression after exposure to ZnO NPs at Sub-MIC level (125 μg/ml). The results of this study are consistent with research that confirmed that PCR technique showed a decrease in the gene expression of gene rsbA in the presence of the nanoinhibitor material, which is zinc oxide nanoparticles ZnONPs and silver nanoparticles AgN.(50). Bacteria use a variety of efflux mechanisms to extrude metal ions beyond cellular boundaries, thus blocking their entry. It has been observed that the exposure of therapeutically relevant bacteria to NPs leads to the upregulation of genes encoding efflux pumps. (51) Reducing the harmful consequences of synthetic processes, the chemicals they include, and the complexes they produce is the main objective. One useful strategy in green nanotechnology is the creation of nanoparticles using different biomaterials. Moreover, utilizing nature’s biological attributes in a range of ways is a great strategy. During the past several years, many materials, such as algae, plants, bacteria, and fungi, have been utilized to create nontoxic, low-cost, and energy-efficient metallic nanoparticles. (52)
Conclusion
Over the past few decades, numerous attempts have been made to develop revolutionary green synthesis techniques. The production of nanomaterials by living organisms holds great promise for application in a variety of fields, including healthcare. Proteus spp. are only a few of the bacterial genera that exhibit the swarming phenomenon and are recognized as having significant pathogenicity. Such bacterial infections caused by swarming germs might be treated. The higher incidence of Proteus species and virulence factors in the present study could be caused by variations in the sanitary methods used. In P. mirabilis treated with ZnO-NPs, the expression of the rsbA gene was dramatically downregulated, whereas ZnO-NPs had no discernible influence on rsbA gene expression. These results imply that by controlling P. mirabilis gene expression, ZnO-NPs may be used as antimicrobial agents.
Acknowledgement
None
Conflict of Interest
There is no conflict of interest
Funding source
There are no funding source.
References
- Al-Mijalli, S.H.S., Shami, A.Y., Al-Salem, R.A. and Alnafisi, N.M . Development of Diagnostic Capabilities for Complications of Bacterial Infection in Diabetic Patients. Review of Diabetic Studies 2022 ; 18(2), pp.135-139.
CrossRef - Al Otraqchi, K.I.B., Darogha, S.N. and Ali, B.A. Serum levels of immunoglobulin and complement in UTI of patients caused by Proteus mirabilis and using AgNPs as antiserum. Cellular and Molecular Biology 2021 ; 67(3), pp.11-23.
CrossRef - De Freitas, C. D . Characterization of swarm-colony development reveals the release of a distinct cell type facilitating dissemination of Vibrio parahaemolyticus (Doctoral dissertation, Philipps-Universität Marburg) 2019.
CrossRef - Abdullah, P.B., Khalid, H.M. and Mero, W.M. Molecular characterization and antibiotic susceptibility of Proteus mirabilis isolated from different clinical specimens in Zakho city, Kurdistan Region, Iraq. Zanco Journal of Pure and Applied Sciences 2022 ; 34(5), pp.198-207.
CrossRef - Cortes-López, H., Juárez-Rodríguez, M., García-Contreras, R., Soto- Hernández, M. and Castillo-Juárez, I. Old Acquaintances in a New Role: Regulation of Bacterial Communication Systems by Fatty Acids. Trends in Quorum Sensing and Quorum Quenching 2020 ; pp.47-57.
CrossRef - Shaba, E.Y.; Jacob, J.O.; Tijani, J.O.; Suleiman, M.A.T. A Critical Review of Synthesis Parameters Affecting the Properties of Zinc Oxide Nanoparticle and Its Application in Wastewater Treatment. Appl. Water Sci 2021 ; 11, 48. [Google Scholar]
CrossRef - Singh, T.A.; Sharma, A.; Tejwan, N.; Ghosh, N.; Das, J.; Sil, P.C. A State of the Art Review on the Synthesis, Antibacterial, Antioxidant, Antidiabetic and Tissue Regeneration Activities of Zinc Oxide Nanoparticles. Adv. Colloid Interface Sci 2021 ; 295, 102495. [Google Scholar]
CrossRef - I. J. Abed , M. E. Ahmed and S. MH AL-Shimmary . “Rosemary Volatile Oil As A Preservative Agent In Some Canned Meat Foods.” Iraqi Journal Of Agricultural Sciences 2021; 52(155-162).
CrossRef - Ahmed, M. E., & al-awadi, a. Q. Enterococcus faecium bacteriocin efflux pump mexa gene and promote skin wound healing in mice. Journal of microbiology, biotechnology and food sciences 2024 ; e10711-e10711.
CrossRef - Ahmed M.E., Al-Awadi A.Q., Abbas A.F. Focus of Synergistic Bacteriocin-Nanoparticles Enhancing Antimicrobial Activity Assay. Microbiological journal 2023; (6). P. 95—104. https://doi.org/10.15407/ microbiolj85.06.095.
CrossRef - Mais E. Ahmed, Khadija Salama. A Comparison Of The Effects Of Lemon Peel -Silver Nanoparticles Versus Brand Toothpastes And Mouthwashes On Staphylococcus spp. Isolated From Teeth Caries. Iraqi Journal Of Science 2020 ; Vol. 61, No. 8, Pp: 1894-1901. Doi: 10.24996/Ijs.2020.61.8.6
CrossRef - Agnieszka, K.-R.; Jesionowski, T. Zinc Oxide—From Synthesis to Application: A Review. Materials 2014; 7, 2833–2881. [Google Scholar]
CrossRef - Sirelkhatim, A.; Mahmud, S.; Seeni, A.; Kaus, N.H.M.; Ann, L.C.; Bakhori, S.K.M.; Hasan, H.; Mohamad, D. Review on Zinc Oxide Nanoparticles: Antibacterial Activity and Toxicity Mechanism. Nano-Micro Lett 2015 ; 7, 219–242. [Google Scholar] [CrossRef][Green Version]
CrossRef - Faiq, N. H., and Ahmed, M., E . Effect of Biosynthesized Zinc oxide Nanoparticles on Phenotypic and Genotypic Biofilm Formation of Proteus mirabilis. Published Online First 2023; https://doi.org/10.21123/bsj.2023.8067
CrossRef - Ahmed, M. E., Q Al-lam, M., & Abd Ali, D. D. M. Evaluation of antimicrobial activity of plants extract against bacterial pathogens isolated from urinary tract infection among males patients. Al-Anbar Medical Journal 2021 ; 17(1), 20-24.
CrossRef - Sayal, R.A., Alkharasani, N.M., Alsadawi, A.A. and Alquraishi, Z.H.O. Molecular study of biofilm and some antibiotic resistance gene in Proteus mirabilis isolated from children with UTI patients in Al-najaf Governorate. Journal of Pharmaceutical Sciences and Research 2018 ; 10(8), pp.1986-1990.
- Hamed, S.M.; Abushanab, K.M.; Elkhatib, W.F. and Ashour, M.SAminoglycoside resistance patterns of certain gram-negative uropathogens recovered from hospitalized Egyptian patients. British Microbiology Research Journal 2013 ; 3(4): 678.
CrossRef - Kathleen Cusick, Yi-Ying Lee,, Brian Youchak and Robert Belas . Perturbation of FliL Interferes with Proteus mirabilis Swarmer Cell Gene Expression and Differentiation . Journal of Bacteriology 2011; p. 437– 447
CrossRef - Tahreer Hadi Saleh , Saba Talib Hashim , Salma Nassrullah Malik , Bahaa Abdullah Laftaah ALRubaii . Down-Regulation of fliL Gene Expression by Ag Nanoparticles and TiO2 Nanoparticles in Pragmatic Clinical Isolates of Proteus mirabilis and Proteus vulgaris from Urinary Tract Infection. Nano Biomed. Eng 2019 ; Vol. 11, Iss. 4
CrossRef - Karpal Singh and Sira Owibingire. Occurrence of Dental Caries among the Adults Attending a Regional Referral Hospital in Tanzania, ournal of Orofacial Research 2014 ; 4(1):30-34
CrossRef - Mais E. Ahmed, Issra S. Mousa, Mohammad M.F Al-Halbosiy and Entsar J. Saheb . Anti-Leishmaniasis Activity of Purified Bacteriocin Staphylococci and Pyocin Isolated from Staphylococcus aureus and Pseudomonas aeruginosa Iraqi Journal of Science 2018; Vol. 59, No.2A, pp: 645-653. DOI:10.24996/ijs.2018.59.2A.2
CrossRef - Ahmed, M.E.; Ahmed, Z.M.; Thamer, A. The Evolutionary Effects Of Bacillin And S-Pyocin Bacteriocin And Their Effects On Propionibacterium Acnes And Fungi. Biochem. Cell. Arch 2022; 20, Supplement 2, pp. 3645-3649
- Balouiri, M., Sadiki, M. and Ibnsouda, S. K. Methods for in vitro evaluating antimicrobial activity: a review. Journal of pharmaceutical analysis 2016 ; 6, 71-79
CrossRef - Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry 1976 ; 72(1-2), pp.248-254.
CrossRef - Ahmed, Mais Emad; Naser, Wasan; Hassoon, Hassanain Abbood The Study Of The Efficacy Of Bacteriocin Isolated From The Genus Salmonella And Its Role In Treating Basra Water Pollution. Samarra Journal Of Pure And Applied Science 2022 ; 4.3: 79-88.
- Muunim, H.H., Al-Mossawei, M.T.and Emad.ahmed, M. The comparative study among the MRSAcin, nisin a and vancomycin, on biofilm formation by methicillin resistance staphylococcus aureus isolated from food sources. International Journal of Drug Delivery Technology 2019 ; 9 (3), pp. 176-181. Doi: 10.25258/ijddt.9.3.31
CrossRef - Koutu, V., Shastri, L. and Malik, M. M. Effect of NaOH concentration on optical properties of zinc oxide nanoparticles. Materials Science-Poland 2016; 34(4): 819-827.
CrossRef - Mohammed LS, Ahmed ME .Effects of ZnO NPS on Streptococcuspyogenes in vivo, Ann Trop Med & Public Health 2020; 23(IIb): S452. DOI: https://DOI:10.36295/ASRO.2020.23228
CrossRef - Ren, S., Yuan, X., Liu, F., Fang, F., Iqbal, H. M., Zahran, S. A. and Bilal, M. Bacteriocin from lacticaseibacillus rhamnosus sp. A5: isolation, purification, characterization, and antibacterial evaluation for sustainable food processing. Sustainability 2022; 14, 9571,
CrossRef - Noor, Faiq and Mais, Ahmed. Inhibitory Effects of Biosynthesized Copper Nanoparticles on Biofilm Formation of Proteus mirabilis Iraqi Journal of Science, 2024, Vol. 65, No.1, pp: 65-78.
CrossRef - Seddiq,S. Zyara,A.M., & Ahmed, M. E. Evaluation the Antimicrobial Action of Kiwifruit Zinc Oxide Nanoparticles Against Staphylococcus aureus Isolated from Cosmetics Tools. BioNanoScience 2023;1-10 https://doi.org/10.1007/s12668-023-01142-
CrossRef - Mohammed LS, Ahmed ME .Effects of ZnO NPS on Streptococcuspyogenes in vivo, Ann Trop Med & Public Health 2020; 23(IIb): S452. DOI: https://DOI:10.36295/ASRO.2020.23228
CrossRef - Erci F, Cakir-Koc R, Yontem M, Torlak E., “Synthesis of biologically active copper oxide nanoparticles as promising novel antibacterial- antibiofilm agents.,” Preparative Biochemistry & Biotechnology., vol. 50, no. 6, pp. 538-548,2020
CrossRef - Thangeeswari, T. George, A. T. and Kumar, A. A. Optical properties and FTIR studies of cobalt doped ZnO nanoparticles by simple solution method. Indian Journal of Science and Technology 2016 ; 9(1)
CrossRef - Diearamane, S.; Lim, Y.; Wong, L.; Lee, PF. Cytotoxic effects of zinc oxide nanoparticles on cyanobacterium Spirulina (Arthrospira) platensis 2018 ; Peer 6, e4682
CrossRef - Soliman M K Y, Abu-Elghait M, Salem S S and Azab M S. Multifunctional properties of silver and gold nanoparticles synthesis by Fusarium pseudonygamai. Springer link 2022.
CrossRef - Gudiña, E.J., Rocha, V., Teixeira, J.A. and Rodrigues, L.R. Antimicrobial and antiadhesive properties of a biosurfactant isolated from Lactobacillus paracasei ssp paracasei A20. Letters in applied microbiology 2010; 50(4), pp.419-424.
CrossRef - Titus, D., Samuel, E.J.J. and Roopan, S.M., Nanoparticle characterization techniques. In Green synthesis, characterization, and applications of nanoparticles 2019; pp. 303-319. Elsevier
CrossRef - Caron. F. Antimicrobial susceptibility testing: a four facets tool for the Clinician. Journal of anti-infection 201214; 168-174 .
- Durgadevi, R., Kaleeshwari, R., Swetha, T.K., Alexpandi, R., Pandian, S.K. and Ravi, A.V. Attenuation of Proteus mirabilis colonization and swarming motility on indwelling urinary catheter by antibiofilm impregnation: an in vitro study. Colloids and Surfaces B: Biointerfaces, 2020; 194, p.111207.
CrossRef - Caron, F. Antimicrobial susceptibility testing: a four facets tool for the clinician. Journal des anti-infectieux 2012; 14, 168-174.
CrossRef - Grasso, G., Zane, D. and Dragone, R. Microbial nanotechnology: challenges and prospects for green biocatalytic synthesis of nanoscale materials for sensoristic and biomedical applications. Nanomaterials 2019; 10(1), p.11.
CrossRef - Little, K., Austerman, J., Zheng, J. and Gibbs, K.A. Cell shape and population migration are distinct steps of Proteus mirabilis swarming that are decoupled on high-percentage agar. Journal of bacteriology 2019; 201(11), pp. e00726-18.
CrossRef - Mohd Yusof, H., Mohamad, R., Zaidan, U.H. and Abdul Rahman, N. A . Microbial synthesis of zinc oxide nanoparticles and their potential application as an antimicrobial agent and a feed supplement in animal industry: a review. Journal of animal science and biotechnology 2019; 10, pp.1-22
CrossRef - Song, Y., Jiang, M., Zhang, H. and Li, R . Zinc oxide nanoparticles alleviate chilling stress in rice (Oryza Sativa L.) by regulating antioxidative system and chilling response transcription factors. Molecules 2021 ; 26(8), p.2196.
CrossRef - AL-Asady, Z.M., AL-Hamdani, A.H. and Hussein, M.A .Study the optical and morphology properties of zinc oxide nanoparticles. In AIP Conference Proceedings 2020; (Vol. 2213, No. 1, p. 020061). AIP Publishing LLC.
CrossRef - Osuntokun, J., Onwudiwe, D.C. and Ebenso, E.E. Green synthesis of ZnO nanoparticles using aqueous Brassica oleracea L. var. italica and the photocatalytic activity. Green chemistry letters and reviews 2019; 12(4), pp.444-457.
CrossRef - Abdelraheem, W.M. and Mohamed, E.S. The effect of zinc oxide nanoparticles on Pseudomonas aeruginosa biofilm formation and virulence genes expression. The Journal of Infection in Developing Countries 2021; 15(06), pp.826-832.
CrossRef - Zhao Vo, N.L.U., Van Nguyen, T.T., Nguyen, T., Nguyen, P.A., Nguyen, V.M., Nguyen, N.H., Tran, V.L., Phan, N.A. and Huynh, Κ.Ρ.Η. Antibacterial shoe insole-coated CuO-ZnO nanocomposite synthesized by the sol-gel technique. Journal of Nanomaterials 2020; pp.1-13.
CrossRef - Ibraheem M. AL-Dulaimy, Hadi R. Rasheed Al-Taai, & Ali Jaffar Saleem. Effect of Silver and Zinc Oxide Nanoparticles on Gene Expression of Some Swarming Genes in Proteus Mirabilis. Central Asian Journal of Medical and Natural Science, 2023 ; 4(3), 11-20.
- Hetta, H.F., Ramadan, Y.N., Al-Harbi, A.I., A. Ahmed, E., Battah, B., Abd Ellah, N.H., Zanetti, S. and Donadu, M.G. Nanotechnology as a Promising Approach to Combat Multidrug Resistant Bacteria: A Comprehensive Review and Future Perspectives. Biomedicines 2023 ; 11(2), p.413.
CrossRef - CHOPRA, Hitesh, et al. Green metallic nanoparticles: biosynthesis to applications. Frontiers in Bioengineering and Biotechnology 2022 ; 10: 548.
CrossRef








