Manuscript accepted on :14-06-2021
Published online on: 24-06-2021
Plagiarism Check: Yes
Reviewed by: Dr. Mohammad Rizki Fadhil Pratama
Second Review by: Dr. Vikas Sharma
Final Approval by: Dr Patorn Piromchai
Hesti L.Wiraswati 1,2* , Fida M. Warganegara3
, Fida M. Warganegara3 , Akhmaloka 3
, Akhmaloka 3 and Muhamad A. Martoprawiro 4
and Muhamad A. Martoprawiro 4
1Oncology and Stem Cell Working Group, Faculty of Medicine, Universitas Padjadjaran, Sumedang 45363, Indonesia
2Department of Biomedical Sciences, Division of Parasitology, Faculty of Medicine, Universitas Padjadjaran, Universitas Padjadjaran, Sumedang 45363, Indonesia
3Department of Chemistry, Institut Teknologi Bandung, Bandung 40132, Indonesia
4Department of Computational Sciences, Institut Teknologi Bandung, Bandung 40132, Indonesia
Corresponding Author E-mail:hesti.lina@unpad.ac.id
DOI : https://dx.doi.org/10.13005/bpj/2170
Abstract
Understanding the metabolism of cytotoxic compounds of quinone family is importance in cancer therapy because they have been successfully explored for their anti-tumor activity. Quinone which form radical semiquinone (by reductase enzymes) to generate Reactive Oxygen Species (ROS) is associated to be anticancer drug candidate. However, molecular mechanism of those compounds to reductase enzymes has not yet clearly understood.This study aimed to understand molecular interaction of quinones to oxidoreductase enzymes such as cytochrome P450 reductase or ubiquinone reductase (NQO1), or apoptosis inducing factor (AIF) which is recently reported as NADH:quinone reductase. In silico approach was applied to find the best affinity of each compound to enzymes. Optimize ligands were employed using Marvin sketch program. Molecular interaction using autodockvina software was built to measure important residues for quinone reduction. Docking analysis showed that generally quinones prefer bound to cytochrome P450 reductase rather than NQO1 or AIF. The number of ring seems affect to the affinity, but not for its functional groups. Residues analysis confirmed that reduction of quinone is NAD(P)H: dependent. The result revealedthat all ligands have high possibility to compete with their redox coupleswhich is needed in its capacity as an anti-cancer drug.
Keywords
Anticancer; Docking; Quinones; Reductase Enzymes; ROS
Download this article as:| Copy the following to cite this article: Wiraswati H. L, Warganegara F. M, Akhmaloka A, Martoprawiro M. A. Molecular Docking Studies of ROS Agent from Quinone Family to Reductase Enzymes:Implication in FindingAnticancer Drug Candidate. Biomed Pharmacol J 2021;14(2). |
| Copy the following to cite this URL: Wiraswati H. L, Warganegara F. M, Akhmaloka A, Martoprawiro M. A. Molecular Docking Studies of ROS Agent from Quinone Family to Reductase Enzymes:Implication in FindingAnticancer Drug Candidate. Biomed Pharmacol J 2021;14(2). Available from: https://bit.ly/2SSATVz |
Introduction
Drugs belonging to the quinone family have been successfully explored for their anti-tumor activity. Understanding the cellular metabolism of their cytotoxic compounds is importance in the field of oncology for finding new therapeutic strategy. Generally, cytotoxicity of quinones is related to its rapid redox cycling which associated with Reactive Oxygen Species (ROS) generation to provoke oxidative stress. Quinones undergo either one-electron reduction to form radical semiquinones or two-electron reduction to form less toxic compound hydroquinones.1,2Those formations may react with oxygen to produce superoxide radicals O2.-. Those formations have a short lifetime in the cells and reactive.3The ROS radical consequences on the occurrence a series of chain reactions of neutral species in the cells or transformation into other types of ROS such as hydrogen peroxide formation. ROS is highly damaging for cells such as vandalized an important macromolecules which consequences on the disruption of their normal functions in the cell. Excessive ROS production will be detrimental to cells by damaging lipid membranes, proteins, and DNA.3–5Normal and cancer cells have different ROS limit level. The researchers reported that cancer cells have an endogenous ROS concentrations greater than normal cells.6,7 Increasing the amount of ROS in cancer cells does not necessarily affect to cell death, due to differences limit of oxidative stress in cancercells. Therefore, it becomes important to study the strong stress agents to kill cancer cells.
The use of compounds that produce ROS as their main mechanism for cell death, are often selected as a drug clinically, although their mechanism to produce ROS has not clearly understood yet. Some quinones are used as chemotherapy, such as mitoxantrone and menadione. Their reduction ability is believed to be responsible in ROS production. Mitoxantrone is anticancer agents that used clinically for the breast cancer, leukemia, lymphoma, and a variety of other malignant tumors treatment.8,9 Menadione has two capacities considered in doing lethal action as an anticancer agent; involved in the redox cycle related to ROS production10,11and menadione has arylation capacity to form conjugation product with thiols proteins in the cell caused a decline in the levels of antioxidants in the cells.11–14Two capacities of menadione has been confirmed by in silico study that showed menadione can interact with NADH domain (correlated with menadione induced ROS production) or FAD domain of AIF (correlated with arylation capacity of menadione).15
The difference of quinone types and its environment affect the oxygen radicals rate, which consequences to cell damage.16 Cytotoxic level of quinones is also affected by enzymatic reducing systems.17,18For instance, P450 reductase is an enzyme that play a major role in one-electron reduction of anticancer compounds. Other enzyme involved in one-electron reduction is ubiquinone oxidoreductase contained in the mitochondria (mitochondrial NADH-dependent ubiquinone oxidoreductase)18or the AIF.19 While, two-electron reduction of quinones generally are catalyzed by NAD(P)H:quinone oxidoreductase 1 (NQO1, DT-diaphorase). This enzyme catalyzes the formation of a neutral compound_hydroquinone, which is less reactive than radical semiquinone.20 The molecular mechanism of quinones to those enzymes has not yet understood. The aim of this study is to understand molecular interaction of quinones to reductase enzymes such as cytochrome P450 reductase, ubiquinone reductase (NQO1), and apoptosis inducing factor (AIF) which is also reported as NADH:quinones reductase.19
Material and Methods
The crystal structure of NQO1, P450 reductase, and Apoptosis Inducing Factor (AIF) with Protein Data Bank (PDB) code subsequently 2F1O, 1AMO, and 3GD4 in complex with its ligands were retrieved from the PDB (http://www.rscb.org/pdb)21,22Three original ligands of each enzyme used in this study; Bishydroxy [2H-1-benzopyran-2-one,1,2-benzopyrone] (C19H12O6, DTC), Nicotinamide Adenine Dinucleotide Phosphate (C21H28N7O17P3, NAP), and Nicotinamide Adenine Dinucleotide (NADH) (NADH).
General Procedure
Protein and ligand preparation
The complexes, heteroatoms and water molecules bound to the receptor molecules were removed from each protein structure. Finally hydrogen atoms were merged to the target receptor molecules. Protein preparation have done using Autodock Vina software.23The ligands were taken from each crystal structure of the enzymes. DTC is ligand of NQO1, NAP is a ligand of P450, and NADH is a ligand of AIF. Quinone compounds were used as ligands are 1,4-benzoquinone, 2-methyl-1,4-benzoquinone, 1,4-naphthoquinone, 2-methyl-1,4-naphthoquinone (menadione), antraquinone, 1,4-dihydroxy-5,8-bis[2-(2-hydroxyethylamino)ethylamino]-anthracene-9,10-dione (mitoxantrone). Ligand structures were sketched using marvin sketch software. Dreiding energy lowest was built to optimize all ligands using marvin sketch software. The optimized ligands were prepared for docking studies.
Docking analysis
The docking analysis of AIF with ligands was carried out by auto dock vina docking software which is most commonly available software. Grid resolution was set to 1 Å, located to NADH binding domain.Docking analysis of AIF-menadione and dreiding energy lowest of menadione were taken from previous report that showed -6.3 kcal/mol and 34.05 kcal/mol respectively.15
Superposition analysis
Superposition of 3D structure of receptors were studied in order to know the folding similarity of both protein, especially on transferase binding domain. This analysis was performed by FAT CAT rigid/flexible-structure alignment.24
Results and discussion
Validation of docking system
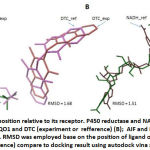
This report begins by showing the validation result of docking program settingin order to confirm the feasibility of system used. Ligand-receptor interaction on crystal structure was used as reference, such as a number of contact residues, the type of contact residues, the number of hydrogen bonds, and Root Mean Standard Deviation (RMSD). This information was retrieved from PDB. All parameters were compared to docking experiment of each ligand-receptor interaction. When the experiment has high similarity with the reference and the value of RMSD ≤ 2, the experimental system will be approved. The result showed that the setup of docking program used in this experiment is good enough in predicting the receptor-ligand interactions. (Table 1, Figure 1)
Table 1: Validation parameters of docking program
| Receptor-Ligand | P450-NAP | NQO1-DTC | AIF-NADH |
| % similarity of contact residues | 86% | 100% | 73% |
| RMSD | 0.97 | 1.68 | 1.51 |
Caption: % similarity of contact residues was measured by identify the type of contact residues of receptor on experiment compare to contact residues of receptor on crystal structure (as reference) RMSD was employed base on the position of ligand on crystal structure (as reference) compare to docking result using autodockvina software.
After validating the docking system, three reductase enzymes as receptor and quinones as ligands were prepared in order to get the lowest energy for interaction. Three dimensional structure of P450 reductase (1AMO), NQO1 (2J1O) and AIF (3GD4) were prepared by removing their original ligand and water molecules from the complexes. (Figure 2) Quinone compounds were prepared as ligand by optimizing the structure using dreiding energy force field. (Figure 2)
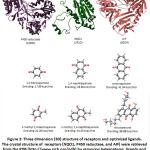 |
Figure 2: Three dimension (3D) structure of receptors and optimized ligands. The crystal structure of receptors (NQO1, P450 reductase, and AIF) were retrieved from the PDB. |
The number of ring affect to quinone’s affinity to enzymes.
Docking results between reductase enzymes and quinones are tabulated in Table 2. Docking analysis showed that anthraquinone have higher affinity of all enzymes compared toother quinones. Affinity energy of anthraquinone<naphtaquinone<benzoquinone. The number of benzene rings seems affect to the affinity, affinity will increase with increasing number of rings. These results seem to be in line with previous studies which revealed that AIF was able to perform the most efficient enzymatic activity against benzoquninone, followed by naphthoquinone and anthraquinone[19]. The lower the affinity of benzoquinone to AIF, with the same rate constant of the enzyme, it will make AIF perform more efficiently.
Table 2: Affinity energy of quinones to P450 reductase, NQO1, and AIF
| Receptor/Ligand | P450 reductase
(kcal/mol) |
NQO1
(kcal/mol) |
AIF
(kcal/mol) |
| 1,4-benzoquinone | -5.2 | -4.3 | -4.8 |
| 2-methyl-1,4-benzoquinone | -5.1 | -4.3 | -4.8 |
| 1,4-naphthoquinone | -7.1 | -5.9 | -6.5 |
| 2-methyl-1,4-naphthoquinone | -6.7 | -5.5 | -6.9 |
| Antraquinone | -8.4 | -6.4 | -8.1 |
| Mitoxantrone | -7.8 | -5.2 | -7.8 |
| NADH | n.d | -6.6 | -9.3 |
| NAP | -8.8 | n.d | n.d |
| DTC | n.d | -7.2 | n.d |
Caption: n.d: not determined
P450 reductase-ligand interaction analysis showed that naphthoquinones and anthraquinones were held by 2 or 3 contact residues more than benzoquinones. (Figure 3) Even, other anthraquinone, mitoxantrone, interacts with 12 contact residues. The number of contact residues may effect to the bigger affinity of mitoxantrone to P450 reductase. While antraquinone, the number of benzene ring appears have a greater contribution (compared to Van der Waals interaction) to its affinity energy value. (Table 3)
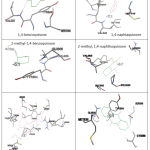 |
Figure 3: Binding pocket of P450 reductase to each quinones |
Table 3: Ligands properties and its contact residues on P450 reductase
| ligands | Contact residues on P450 reductase | Water Solubility, pH 7.4 | Polar Surface Area (PSA) |
| 1,4-benzoquinone | ser596, val605, tyr604 | High (higher than 0.06 mg/ml) | 34.14 |
| 2-methyl-1,4-benzoquinone | val605, tyr604 | High (higher than 0.06 mg/ml) | 34.14 |
| 1,4-naphthaquinone | gly565, ser596, val605, tyr604, met636 | High (higher than 0.06 mg/ml) | 34.14 |
| 2-methyl-1,4-naphthaquinone | val605, tyr604, ser596, gly565, met636 | High (lower than 0.01 mg/ml) | 34.14 |
| anthraquinone | cys566, ser596, tyr604, gln606, met636 | Low | 34.14 |
| Mitoxantrone | arg298, leu300, pro533, gly534, tyr564, cys566, asp572,ser596, tyr604, val605, met636, asn635 | High (higher than 0.06 mg/ml) | 152.11 |
Caption: Contact residues of enzyme were performed as results of docking analysis. Ligand properties parameters (water solubility and polar surface area) were measured using marvin sketch software.
The presence of methyl group on each quinone, seems not affect to the affinity (Table 2, Figure 2). Other factors such as water solubility and Polar Surface area (PSA) do not appear to contribute to the affinity of ligand to P450. (Table 3) Docking analysis of quinones to other reductase enzymes (NQO1 and AIF) indicate the similar result.
Binding energy analysis showed that generally, quinones prefere to bind to cytochrome P450 reductase rather than to NQO1 or AIF.
In general, quinones prefer bound to P450 reductase rather than to AIF or NQO1 (Table 2). It may be affected by reduction potential value of each quinones. Benzoquinone has reduction potential higher followed by naphthoquinone then antraquinone.19Reduction process is often achieved stepwise via one-electron reduction. The intermediate product (semiquinone molecules) is a free radical. The reduction potentials of such one-electron couples (quinone/semiquinone) are value in predicting the direction of many free-radical reactions. Meaning, quinone which has higher reduction potential’s value tends to interact with enzymes that facilitate one-electron reduction. It can explain why all ligands (benzoquinone or naphthaquinone or anthraquinone) have better affinity to P450 reductase then followed by AIF, rather than NQO1. This result confirmed the ability of AIF to catalyze one-electron reduction.19Even, 2-methyl-1,4-naphthoquinone showed different phenomenon. Here, 2-methyl-1,4-naphthoquinone interacts to AIF better than interacts to P450 reductase.Analysis of ligand-receptor interaction revealed also that there is a residue that consistent interact to each quinones. Tyr604 support quinone interaction using Van der Waals interaction. It appears that Tyr604 pose 90o to central ring on each quinone. Thus, Tyr604 residues may acts as recognizer residue the target compound of P450 reductase. (Table 3)Here also showed that the energy values of quinones-AIF close to quinones-P450 reductase (rather than to quinones-NQO1). The structural alignment of three enzymes was applied to explain this situation. FAT CAT program revealed that the folding of AIF close to P450 reductase rather than NQO1. (Figure 4)
 |
Figure 4: Folding alignment between AIF (3GD4) to P450 reductase (1AMO) or NQO1 (2F1O). AIF has 65% folding similarity to P450 reductase (A). AIF has 35% folding similarity to NQO1 (B). |
Residues analysis confirmed that reduction of quinone is NAD(P)H dependent
Several contact residues to the binding of NAD(P)H as redox couple of quinones, were used to do analysis NAD(P)H-dependent on quinones-enzymes interaction. (Table 2) Here, 2-methyl-1,4-benzoquinone was used as a ligand model in order to understand the relationship between ligand to its redox couple on each reductase enzymes. Receptor-ligand interaction analysis showed that contact residues of target ligand are fit with contact residues of its couple redox. (Figure 5)This result revealed that the target ligand has high possibility to compete with redox couples, although it requires more support factor to overcome the obstacle threshold (ex. concentration), given the affinity energy of 2-methyl-1,4-benzoquinone is under their redox partner.These results confirmed that AIF can act as NADH:quinone reductase.19
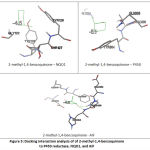 |
Figure 5: Docking interaction analysis of of 2-methyl-1,4-benzoquinone to P450 reductase, NQO1, and AIF |
Conclusion
Exploration of compounds that has capacity to kill cancer cells is challenge. One is to find potential anti-cancer agent which is associated with its ability to stimulate oxidative stress through generate ROS production. Given that cancer cells have higher threshold of ROS than normal cells, finding the ROS agent is absolutely necessary. One type of compound that is widely explored for its anticancer abilities are quinone family. Quinone have reduction capacity to form radical semiquinone or the less-reactive compound, hydroquinone. These reductions is associated with reducing enzymes including P450 reductase, NQO1, and AIF. This report revealed that in general quinones prefer bound to cytochrome P450 reductase rather than NQO1 or AIF. Several factors indicated influence to this process; the amount of quinone ring and reduction potential value. Functional groups of the ligand and Polar Surface area (PSA) do not appear to contribute to the affinity. The lower affinity were found for quinones-NQO1 interaction compared to quinones-AIF or Quinones-P450 reductase interaction. Residues analysis confirmed that reduction of quinone is NAD(P)H:dependent. Further analysis revealed that all quinones have high possibility to compete with their redox couples, although it requires more support factor to overcome the obstacle threshold. Finally, all describes that all quinones on this study have the potential as anticancer.
Acknowledgment
The authors would like to humbly acknowledge The Indonesian Ministry of Research, Technology and Higher Education (Ristekdikti) for financial support during the research.
Conflict of Interest
No potential conflict of interest was reported by the authors.
Funding Source
The present study was funded by The Indonesian Ministry of Research, Technology and Higher Education (Ristekdikti).
References
- O’Brien, P. J. Molecular mechanisms of quinone cytotoxicity. Chem. Interact.80, 1–41 (1991).
CrossRef - Morgan WA1, Hartley JA, C. G. Quinone-induced DNA single strand breaks in rat hepatocytes and human chronic myelogenous leukaemic K562 cells. Biochem Pharmacol.44 (2), 215–21 (1992).
CrossRef - Pelicano Helena, Carney Dennis, H. P. ROS stress in cancer cells and therapeutic implications. Drug Resist. Updat.7, 97–110 (2004).
CrossRef - R., S. E. Metal ion-catalyzed oxidation of proteins: biochemical mechanism and biological consequences. Free Radic Biol Med9, 315–25 (1990).
CrossRef - Hensley Kenneth, Robinson Kent A., Gabbita S. Prasad, Salsman S., F. R. A. Reactive oxygen species, cell signaling, and cell injury. Free Radic Biol Med28, 1956–62 (2000).
CrossRef - Hileman E.O., Liu J., Albitar M., Keating M.J., H. P. Intrinsic oxidative stress in cancer cells: a biochemical basis for therapeutic selectivity. Cancer Chemother Pharmacol53, 209–19 (2004).
CrossRef - Behrend L., Henderson G., Z. R. M. Reactive oxygen species in oncogenic transformation. Biochem. Soc. Trans.31, 1441–4 (2003).
CrossRef - Ehninger Gerhard, Schuler Ulrich, Proksch Barbara, Zeller Klaus-Peter, B. J. Pharmacokinetics and Metabolism of Mitoxantrone A Review. Clin. Pharmacokinet.18, 365–80 (1990).
CrossRef - Faulds Diana, Balfour Julia A., Chrisp Paul, L. H. D. Mitoxantrone A Review of its Pharmacodynamic and Pharmacokinetic Properties, and Therapeutic Potential in the Chemotherapy of Cancer. Drugs41, 400–449 (1991).
CrossRef - Daugas, E. et al. Apoptosis-inducing factor (AIF): A ubiquitous mitochondrial oxidoreductase involved in apoptosis. FEBS Lett.476, 118–123 (2000).
CrossRef - Wiraswati, H. L. et al. Apoptosis inducing factor (AIF) mediates lethal redox stress induced by menadione. Oncotarget7, 76496–76507 (2016).
CrossRef - Di Monte Donato, Ross David, Bellomo Giorgio, Eklow Lena, O. S. Alterations in intracellular thiol homeostasis during the metabolism of menadione by isolated rat hepatocytes. Biochem Biophys235, 334–342 (1984).
CrossRef - Wang, X. et al. Mechanism of arylating quinone toxicity involving Michael adduct formation and induction of endoplasmic reticulum stress. Proc. Natl. Acad. Sci.103, 3604–3609 (2006).
CrossRef - Scott, G. Vitamin K3 (menadione) induced oncosis associated with keratin 8 phosphorylation and histone H3 arylation. Mol. Pharmacol.68, 606–615 (2005).
CrossRef - Wiraswati, H. L., Martoprawiro, M. A. & Warganegara, F. M. Apoptosis Inducing Factor ( AIF ) Stabilizes Menadione- Conjugate Product in Programmed Cell Death. 10, 237–245 (2017).
CrossRef - Rockwell S., Sartorelli A. C., Tomasz M., K. K. A. Cellular pharmacology of quinone bioreductive alkylating agents. Cancer Metastasis Rev.12, 165–76 (1993).
CrossRef - Gibson N.W., Hartley J.A, Siegel D., R. D. Relationship between DT-diaphorase-mediated metabolism of a series of aziridinylbenzoquinones and DNA damage and cytotoxicity. Mol. Pharmacol.42, 531–36 (1992).
- Mustofa, H. N., Wiraswati, H. L., Ekawardhani, S., Fianza, P. I. & Windria, S. Anticancer Activities of Saponins and Quinones Group through Oxidative Stress and Glycolysis Inhibition via in Silico Studies. Biomed. Pharmacol. J.13, 999–1010 (2020).
CrossRef - Iyanagi Takashi, Y. I. One-electron-transfer reactions in biochemical systems V. Difference in the mechanism of quinone reduction by the NADH dehydrogenase and the NAD(P)H dehydrogenase (DT-diaphorase). Biochim. Biophys. Acta216, 282–94 (1970).
CrossRef - Misevičien, L., Anusevičius, Ž., Šarlauskas, J., Sevrioukova, I. F. & Čnas, N. Redox reactions of the FAD-containing apoptosis-inducing factor (AIF) with quinoidal xenobiotics: A mechanistic study. Arch. Biochem. Biophys.512, 183–189 (2011).
CrossRef - L., G. P. The role of NAD(P)H oxidoreductase (DT-Diaphorase) in the bioactivation of quinone-containing antitumor agents. Free Radic. Biol. Med.29, 263–75 (2000).
CrossRef - Wang, M. et al. Three-dimensional structure of NADPH-cytochrome P450 reductase: Prototype for FMN- and FAD-containing enzymes. Proc. Natl. Acad. Sci.94, 8411–8416 (1997).
CrossRef - Asher Gad, Dym Orlt, Tsvetkov Peter, Adler Julia, S. Y. The crystal structure of NAD(P)H quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol. Biochemistry45, 6372–78 (2006).
CrossRef - Trott Oleg, O. A. J. autodock vina:improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem.31, 455–61 (2010).
CrossRef - Ye Y., G. A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics2, 246–55 (2003).
CrossRef