Sangeetha M1 , Saranya T S1
, Saranya T S1 , Sathianarayanan S1
, Sathianarayanan S1 , Hima Vyshnavi A M2
, Hima Vyshnavi A M2 and P K Krishnan Namboori2*
and P K Krishnan Namboori2*
1Department of Pharmaceutical Chemistry and Analysis, Amrita School of Pharmacy, Amrita Vishwa Vidyapeetham, Amrita Institute of Medical Sciences, AIMS Health Science Campus, AIMS Ponekara P O, Kochi-682041.
2Computational Chemistry Group (CCG), Amrita Molecular Modeling and Synthesis Research lab,Amrita School of Engineering, Coimbatore, Amrita Vishwa Vidyapeetham, India.
Corresponding Author E-mail : n_krishnan@cb.amrita.edu
DOI : https://dx.doi.org/10.13005/bpj/1933
Abstract
The antibiotic resistance is overwhelming at an alarming rate. These ‘resistant bacteria’ affect millions of people in their health care practices worldwide. The misuse of antibiotics and the overuse of antibiotics in humans, as well as animals may lead to accelerating the process. The ‘Methicillin-Resistant Staphylococcus Aureus (MRSA) is the most common antibiotic-resistant bacterium in humans identified at present and is obtained through the mecA gene transcription. In spite of all modern strategies available to minimize the MRSA resistance, a new effective antimicrobial treatment is necessary to control infections. In this study, we designed and developed a new potential flavonoid moiety for MRSA therapy. Various computational methods are used for identifying the best compound for the treatment for this therapy including docking studies, ADEMTox analysis and the characterization of the respective mechanisms. The target protein molecules have been designed through homology modelling and potential flavonoid molecules have been suggested from PubChem database. Hesperetin ((S)-5,7-dihydroxy-2-(3-hydroxy-4-methoxyphenyl)chroman-4-one), has been identified as a potential flavonoid moiety suitable for MRSA.
Keywords
Antibiotic Resistance; Flavonoids; Hesperetin; Homology Modelling; MRSA
Download this article as:| Copy the following to cite this article: Sangeetha M, Saranya T S, Sathianarayanan S, Vyshnavi A. M. H, Namboori P. K. K. Design and Development of Potential Flavonoid Moiety for Pbp2a Inhibition for Mrsa Therapy-A Computational Technique. Biomed Pharmacol J 2020;13(2). |
| Copy the following to cite this URL: Sangeetha M, Saranya T S, Sathianarayanan S, Vyshnavi A. M. H, Namboori P. K. K. Design and Development of Potential Flavonoid Moiety for Pbp2a Inhibition for Mrsa Therapy-A Computational Technique. Biomed Pharmacol J 2020;13(2). Available from: https://bit.ly/3b92XH3 |
Introduction
Methicillin-Resistant Staphylococcus aureus is a commonly known bacterium that leads to several infections in humans. It is the major cause of multidrug-resistant infections with significant morbidity and mortality. Based on the reports of ‘Centers for disease control and prevention (CDC)’ in 2011, 80,000 invasive infections and 11,000 deaths were reported in the United States. An estimated 2 billion people carry staphylococcus aureus; about 53 million people are thought to carry MRSA. [1].
The common mechanisms used by bacteria to minimize the effects of antibiotics include changes in the antibiotic molecule, reduced antibiotic penetration and efflux, changes in the target site and resistance due to global cell adaptations, antibiotic sequestration, etc. The most important mechanism behind the antibiotic resistance is often acquired by the transfer of resistance-conferring genes between bacteria facilitated by a conjugative plasmid [2,3].
Staphylococcus aureus is a gram-positive bacterium that commonly seen inside the nose and on human skin. These bacteria are resistant to numerous antibiotics. Methicillin is an antibiotic widely uses against Staphylococcus aureus. Once the bacteria develop a self-resistance to methicillin, the resistant bacteria is known as MRSA. Mainly all antibiotics are resistant to all microorganisms. The mecA gene, which is encoded with penicillin-binding protein (PBP2a) and is with reduced affinity for β-lactam antibiotics, acquires methicillin resistance. The mecA gene, found in bacterial cells, is responsible for developing resistance to penicillin related antibiotics.
In the ‘mec operator’ is keeping mecA, mecI, mecRI and mecR2 [4], out of which, the mecA gene, encoding ‘penicillin-binding protein 2a (PBP2a)’, acquires Methicillin-Resistant Staphylococcus Aureus (MRSA) infection. The inhibition of penicillin-binding protein 2a (PBP2a) has been identified as a potential therapeutic technique to overcome MRSA. This could be possible by using the antibiotic along with a supporting natural product such as flavonoid, which would inhibit the PBP2a activity [5].
The pathophysiology of methicillin resistance includes the transcription of the mecA gene into the cell wall producing resistance and leading into the degradation of cell membrane. So, the transcription of the mecA gene will produce resistance against the drug.
MRSA is the main carrier of the mecA gene, which can undergo horizontal gene transfer into the host species [ 6] through a mobile genetic element, Staphylococcal Cassette Chromosome (SCC) mec . The protein targets of PBP2a of Staphylococcus aureus does not have any crystalized structure in the database. However, the protein models can be generated through ‘homology modeling’ by taking one of the biosynthesized proteins as the template [ 7, 8]. The interaction between the target protein models and potential drug molecules can be studied through ‘docking studies’ [9-13]. Further, pharmacokinetic and pharmacodynamic parameters can also be predicted to make screening of the molecules identified.
In this work, the structural protein model molecules of the mecA gene and their interactions with selected flavonoids have been carried out. This would provide an insight into finding a suitable and potential flavonoid moiety for the treatment of MRSA infection.
Materials and Methods
The mecA gene protein sequence has been identified using BLAST search and is further subject to homology modelling using ‘Swiss Model’ by taking 1MWT as the template [14-16]. The ‘protein model’ characterization has been carried out to study the quality of the models. The flavonoid molecules have been selected from PubChem as the ligand molecules.
The phytoconstituents like Epicatechin gallate, Dihydroquercetin, Fisetin, Myricetin, Farrerol, Peonidin, Quercetol, Hesperetin, Luteolin, Isorhamnetin, Epicatechin, Morin, Phloretin, Naringenin, Catechin, Kaempferol, Shogaol, Glycitein, Strobopinin, Apigenin, Genistein, Daidzein, Ellagic acid, Isoquercetin, etc. were selected. The model protein is further docked with the selected flavonoid molecules [17-18]. The drug-likeness has been further checked through ADMETox studies to identify the most suitable flavonoid to be used along with methicillin to arrest MRSA.
Results and Discussions
The protein models have been designed and developed using homology modelling using the biosynthetic protein, 1MWT as the template. The model protein showed, 96% sequence similarity towards the template protein. The structure of 1MWT and the model protein are shown in Fig 1.
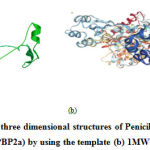 |
Figure 1: (a) Predicted three dimensional structures of Penicillin-Binding Protein 2a |
An interaction study has been carried out with the ligand molecules and the modelled protein structure. It has been observed that the amino acids GLU33, SER 76, ARG 36 and Val 64 were found to be interacting with the ligand molecules Fig 2 as expected through the mechanism of MRSA [19]. The phenolic hydroxyl groups in position 7 and the -OH group at position 3 on the C-ring of Hesperetin are found to be interacting site of the flavonoid moiety.
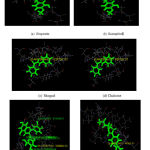 |
Figure 2: Docking interaction of selected phytoconstituents in modeled protein. |
Among the flavonoid molecules used for the analysis, Hesperetin (Fig 3) is found to be most interacting with the model protein. In fact, this molecule has been identified as more interacting than the standard drug, linezolid used as the control molecule Table 1.
Table 1: Phytochemicals and their Interaction Energy and Docking score
| MOLECULE | DOCKING SCORE
(kcal/mol) |
DOCKING INTERACTION (kcal/mol) |
| Chalcone
Shogaol Hesperetin Isoquercetin Kaempferol Linezolid – Std |
22.298
31.733 34.041 15.027 32.359 28.623 |
32.268
42.379 42.560 50.301 37.686 41.820 |
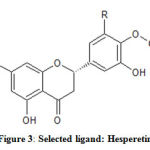 |
Figure 3: Selected ligand: Hesperetin |
The drug-likeness studies suggest Kaempferol and Hesperetin as potential flavonoids with favourable number of ‘hydrogen bonding acceptors and donors Table 2.
Table 2: Drug-likeness prediction through Biovia Discovery Studio software
| Sl.No.
|
Ligands
|
AlogP
|
MW
|
No. of HBA | No. of HBD |
| 1
2 3 4 5 |
Chalcone
Shogaol Hesperetin Isoquercetin Kaempferol |
3.701
4.717 2.357 -0.3 1.872 |
208.255
276.371 302.279 464.376 286.236 |
1
3 6 12 6 |
0
1 3 8 4 |
All the molecules are keeping good absorption excepting isoquercetin. Hesperain and Kaempferol showed good solubility and less blood-brain barrier penetration. However, these molecules are expected to be more hepatotoxic than the control drug (Table 3).
Table 3: ADMET study of selected ligands
| Sl. No. | Ligands | ADMET Solubility log(sw) – (S) | ADMET BBB ratio
(R) |
ADMET Hepatotoxic
Probability |
ADMET Absorption level (HIA) | ADMET AlogP 98 |
| 1
2 3 4 5 |
Chalcone
Shogaol Hesperetin Isoquercetin Kaempferol |
-6.00 S< -4.00
-6.00<S<-4.00 -4.00<S<-2.00 -6.00<S<-4.00 -4.00<S< -2.00 |
R>5:1
1:1<R< 5:1 R<0.3:1 undefined R<0.3:1 |
<0.5
<0.5 >0.5 <0.5 >0.5 |
<6.12
<6.12 <6.12 >7.00 <6.12 |
3.702
4.717 2.357 -0.3 1.872 |
Conclusion
The extensive research into the interactions of flavonoid compounds with the penicillin-binding proteins of many species has shown the variety of the sensitivity of individual PBPs. The ‘penicillin-binding proteins (PBP)’ can be potential drug targets. The protein models have been generated through homology modelling and are used for studying the possibility of using flavonoids along with Methicillin for controlling MRSA. While studying the interaction between these molecules and the model protein molecules, Hesperetin is found to be most favourable potential molecule. The interacting aminoacids present in the protein models are found to be in the expected binding site. Hence, Hesperetin ((S)-5,7-dihydroxy-2-(3-hydroxy-4-methoxyphenyl)chroman-4-one) can be tried as a flavonoid to be used along with methicillin for MRSA therapy subject to further invitro-invivo evaluations.
Acknowledgement
The author thankful to Department of Pharmaceutical Chemistry and Analysis, Amrita School of Pharmacy, Amrita Molecular Modeling And Synthesis (AMMAS) resaech lab for their support in completing this work.
Complaince with Ethical Standards
Not Available
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- Sangeetha Joshi, Pallab Ray, Vikas Manchanda (2013),Methicillin resistant Staphylococcus aureus (MRSA) in India: Prevalence & susceptibility pattern, Indian J Med Res.137(2): 363-369.
- Elizabeth Peterson and Parjit Kaur (2018), Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria and Clinical Pathogens, Front.Microbiol.9:2928, https://doi.org/10.3389/fmicb.2018.02928.
- M. Rozwandowicz, M. S. M. Brouwer, J. Fischer, J.A. Wagenaar (2018), Plasmids carrying antimicrobial resistance genes inEnterobacteriaceae, J.Antimicrob.Chemother. 73:1121-1137, https://doi.org/10.1093/jac/dkx488.
- Sahra Kirmusaoglu (2017), MRSA and MSSA: The Mechanism of Methicillin Resistance and the Influence of Methicillin Resistance on Biofilm Phenotype of Staphylococcus aureus, INTECH open science, DOI: 10.5772/65452.
- Santiago C, Pang EL, Lim KH, Loh HS, Ting KN. Inhibition of penicillin-binding protein 2a (PBP2a) in methicillin resistant Staphylococcus aureus (MRSA) by combination of ampicillin and a bioactive fraction from Duabanga grandiflora.BMC Complement Altern Med. 2015:15:178
- Sharon J. Peacock and Gavin K. Paterson (2015), Mechanisms of Methicillin Resistance Staphylococcus aureus, Annu. Rev. Biochem.84:577-601, https://doi.org/10.1146/annurev-biochem-060614-034516.
- Rekha Patil, Shrikanth R K, Mohan Reddy K, G. R. Naik1. Drug Interface Residues Of Penicillin Binding Protein 2a: An Insilico Structural Analysis And Docking Studies For Potential Drugs On MRSA. Research. RJLBPCS. 2018. 4(4) Page No.594
- Andrew Waterhouse, Martino Bertoni, Stefan Bienert, (2018), SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Research, Vol.46, 2, https://doi.org/10.1093/nar/gky427.
- A.M. Vijesh, Arun M. Isloor, Sandeep Telkar, T. Arulmoli, Hoong-Kun Fun (2013), Molecular docking studies of some new imidazole derivatives for antimicrobial properties, Arabian Journal of Chemistry6, 197-204, https://doi.org/10.1016/j.arabjc.2011.10.007.
- P. M. Iyer, P. Kumar, S., Shanmugam, K., and P. K. Krishnan Namboori, “Retrometabolic approach for designing personalized anti- cancer drug molecules for controlling breast cancer resulted by BRCA1 mutations”, Current Pharmacogenomics and Personalized Medicine (Formerly Current Pharmacogenomics), vol. 14, no. 1, pp. 56 – 64, 2016.
- Preethi M Iyer, Karthikeyan S, Sanjay KUMAR Palayat, Krishnan Namboori. Comprehensive strategy for the design of precision drugs and identification of genetic signature behind proneness of the disease-a pharmacogenomic approach. Functional & Integrative Genomics. 2017:17(4).
- P. M Iyer, Kumar P Sanjay, Karthikeyan S and P. K. Krishnan Namboori, “Brca1 responsiveness towards breast cancer-a population-wise pharmacogenomic analysis”, International Journal of Pharmacy and Pharmaceutical Sciences, 2016:8,267-270.
- C. P. Anju, Anusooya N. J, Deeasree M., Deepak O. M. and P. K. Krishnan Namboori, “De Novo designing of HDAC inhibitors in cancer therapy”, International Journal of Pharmaceutical Science and Health Care. 2012:2,59–66.
- Mark Johnson, Irena Zaretskaya (2008), NCBI BLAST: a better web interface, Nucleic Acids Research, vol.36, W5-W9, doi:10.1093/nar/gkn201.
- C. S Vasavi, Saptharshi, R. Radhika Devi, Lakshmi Anand, Megha. P. Varma and P.K Krishnan Namboori(2010), Homology Modeling and Protein Ligand Interaction to Identify Potential Inhibitor for E1 Protein of Chikungunya, CCIS 101, pp. 510-513. https://doi.org/10.1007/978-3-642-15766-0_85.
- William R. Pearson (2013), An introduction to Sequence Similarity (“Homology”) Searching, Curr. Protoc. Bioinformatics,42:3.1.1-3.1.8.https://doi.org/10.1002/0471250953.bi0301s42.
- Xuan-yu Meng, Hong-Xing Zhang (2011), Molecular Docking: A powerful approach for structure-based drug discovery, Curr comput Aided Drug Des, 7(2):146-157.https://doi.org/10.2174/157340911795677602.
- Shefrin.S, Manakadan A.A, Saranya T.S (2018), A computational study of anticancer activity of curcumin derivatives using In silico drug designing and Molecular docking tools, Asian. Journal of Chemistry, Vol.30, PP. 1335-1339.https://doi.org/10.14233/ajchem.2018.21239.
- Sahra Kirmusaoglu (2017), MRSA and MSSA: The Mechanism of Methicillin Resistance and the Influence of Methicillin Resistance on Biofilm Phenotype of Staphylococcus aureus, INTECH open science. Ann. Rev. Biochem. 2015. 84:577-601.








