Manuscript accepted on :December 19, 2016
Published online on: --
Plagiarism Check: Yes
Mohammad Darvishi
Infectious Diseases and Tropical Medicine Research Center (IDTMRC), AJA University of Medical Sciences, Tehran, Iran.
Corresponding Author Email : mohammaddarvishi@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1048
Abstract
Natural inherent of Acinetobacter baumanni to survive in hard conditions in surfaces and its ability to resist against commonly used antibiotics in hospitals caused it to be one of the most prevalent cause of hospital infections. The present study was carried out to research the prevalence, antibiotic resistance pattern and distribution of virulence genes in the A. baumannii strains of various infections of immunosuppressive patients. One-hundred and fifty samples were collected and cultured. Their positive results were subjected to disk diffusion and PCR. Of all 150 samples studied, 20 samples (13.33%) were infected with A. baumannii. Wound infections had the highest prevalence of A. baumannii (16%). Csga (70%) and cnf1 (50%) were the most commonly detected virulence genes. A. baumannii strains showed the highest levels of resistance against ampicillin (100%), tetracycline (95%), gentamycin (75%) and cephalexin (60%), while lowest against imipenem (5%) and ceftriaxone (35%). Statistically significant difference was seen between the type of samples and prevalence of A. baumannii, prevalence of antibiotic resistance and also distribution of virulence genes (P < 0.05). Quick determination of infections caused by A. baumanni and its treatment with imipenem can decrease the risk of A. baumanni’s infections.
Keywords
Acinetobacter baumanni;Virulence genes;Antibiotic resistance pattern;Immunosuppressive patients;Clinical infections
Download this article as:| Copy the following to cite this article: Darvishi M. Virulence Factors Profile and Antimicrobial Resistance of Acinetobacter baumannii Strains Isolated from Various Infections Recovered from Immunosuppressive Patients. Biomed Pharmacol J 2016;9(3). |
| Copy the following to cite this URL: Darvishi M. Virulence Factors Profile and Antimicrobial Resistance of Acinetobacter baumannii Strains Isolated from Various Infections Recovered from Immunosuppressive Patients. Biomed Pharmacol J 2016;9(3). Available from: http://biomedpharmajournal.org/?p=11737 |
Introduction
Healthcare-associated and hospital-acquired infections (HAIs) are common cause of mortality and morbidity al-around the world. Pathogenic bacteria are the most important causes of HAIs. Among all of the, Acinetobacter baumannii is one of the most prevalent cause of infections in the hospital environment (1, 2).
Acinetobacter species are aerobic gram-negative bacilli that can survive for prolonged periods in the environment and on the hands of healthcare workers (1-3). Furthermore, Acinetobacter infections have become increasingly difficult to treat because of the emergence of strains resistant to various types of antibiotics including cephalosporins, quinolones, sulfonamides, macrolides, aminoglycosides, fluoroquinolones and tetracycline (4, 5). These multidrug-resistant (MDR) strains are responsible for causing various types of infections including endocarditis, wound, skin and soft tissue infections, meningitis, septicemia, pneumonia and respiratory and urinary tract infections (RI and UTIs) (1-3).
Pathogenesis of diseases caused by A. baumannii is derived from the presence of latent virulence genes (6, 7). Some of the most significant virulence genes of the A. baumannii strains of human clinical infections are colicin V production (cvaC), curli fibers (csg), siderophores like aerobactin (iutA) and cytotoxic necrotizing factor (cnf) (6, 7). Detection of latent virulence genes in the clinical isolates of A. baumannii has some great epidemiological outcomes help practitioners to control dissemination of infectious diseases caused by this bacterium.
Up to now, there were no well-conducted previously published data about the prevalence and epidemiology of A. baumanni strains in human clinical samples in Iran. Therefore, the present investigation was done in order to study the prevalence, distribution of virulence genes and antibiotic resistance pattern of A. baumannii strains isolated from various types of infections recovered from immunosuppressive hospitalized patients.
Materials and methods
Samples and Acinetobacter baumannii isolation
From January 2015 to April 2016, a total of 150 infectious samples including wound (n=50), respiratory (n=40) and urine (n=60) samples were collected from immunosuppressive patients hospitalized in hospitals and health care centers of Iran. Samples were collected from less than 70 years old hospitalized patients. Samples were immediately transferred to the laboratory in cooler with ice packs.
Samples were inoculated on to blood agar (Merck, Germany) and MacConkey agar (Merck, Germany) and incubated aerobically at 37°C for 24 hours. Non-hemolytic, opaque and creamy colonies on blood agar and nonlactose fermenting colonies on MacConkey agar were further sub-cultured on MacConkey agar and incubated for another 24 hours at 37ºC to obtained pure colonies. The isolated organisms were identified based on colonial and microscopic characteristics and various biochemical tests according to standard laboratory methods (8). Further identification of isolates was done using Gram stain, oxidase test and API 20NE identification strip (Biomérieux, Marcy l’Etoile, France).
Antimicrobial susceptibility testing
Pattern of antimicrobial resistance was studied using the simple disk diffusion technique. The Mueller–Hinton agar (Merck, Germany) medium was used for this purpose. Antibiotic resistance of A. baumannii strains against commonly used antibiotics was determined using the instruction of Clinical and Laboratory Standards Institute guidelines (9). Susceptibility of A. baumannii strains were tested against levofloxacillin (5 µg/disk), ampicillin (10 u/disk), imipenem (30 u/disk), gentamycin (10 µg/disk), cephalothin (30 µg/disk), cephalexin (10 µg/disk), tetracycline (30 µg/disk), trimethoprim/sulfamethoxazole (25 µg/disk) and ceftriaxone (30 µg/disk) antibiotic agents (Oxoid, UK). All of the inoculated plates were aerobically incubated at 37 °C for 18-24 h in an aerobic atmosphere. Results were interpreted based on the instruction provided by CLSI (2012) (9). In all reactions, the A. baumannii ATCC 19605 was used as quality control bacterium.
DNA extraction from the Acinetobacter baumannii isolates
A single colony of A. baumannii was inoculated on 5 ml of nutrient broth and incubated over night at 37 ºC. Genomic DNA was extracted from the bacterial colony using the genomic DNA extraction kit (Fermentas, Germany) according to the manufacture instruction. The DNA concentration has been determined by measuring absorbance of the sample at 260 nm using spectrophotometer (10).
PCR-based detection of virulence genes
Table 1 indicates list of primers and PCR program used for detection of virulence factors (11). The DNA was amplified in a programmable thermal cycler (Eppendorf, Mastercycler® 5330, Eppendorf-Netheler-Hinz GmbH, Hamburg, Germany). Fifteen microliters of PCR products were resolved on a 1.5% agarose gel containing 0.5 mg/ml of SYBR Green in Tris–borate–EDTA buffer at 90 V for 40 min, also using suitable molecular weight markers. The products were examined under ultraviolet illumination. A. baumannii ATCC 17978 and A. baumannii ATCC 19606 and rough strains purchased from the Pasteur Institute (Tehran, Iran) were used as positive controls and distilled water (D.W, Merck, Germany) was used as a negative control.
Statistical analysis
Statistical analysis was performed using SPSS/21.0 software (SPSS Inc., Chicago, IL). The chi-square test and Fisher’s exact 2-tailed test analysis were performed in this study. Statistical significance was regarded at a P value < 0.05.
Table 1: Primer sequence and PCR conditions used for detection of virulence genes in the A. baumannii isolates of various types of infections.
| Gene target | Primer sequence (5′-3′)* | PCR product (bp) | PCR Volume (50µL) | PCR programs |
| cnf1 | F: AAGATGGAGTTTCCTATGCAGGAG
R: CATTCAGAGTCCTGCCCTCATTATT |
498 | 5 µL PCR buffer 10X
1.5 mM Mgcl2 200 µM dNTP (Fermentas) 0.5 µM of each primers F & R 1.25 U Taq DNA polymerase (Fermentas) 2.5 µL DNA template |
1 cycle:
95 0C ———— 4 min. 30 cycle: 95 0C ———— 50 s 58 0C ———— 60 s 72 0C ———— 45 s 1 cycle: 72 0C ———— 8 min
|
| csgA | F: ACTCTGACTTGACTATTACC
R: AGATGCAGTCTGGTCAAC |
200 | ||
| cvaC | F: CACACACAAACGGGAGCTGTT
R: CTTCCCGCAGCATAGTTCCAT |
680 | ||
| iutA | F: GGCTGGACATCATGGGAACTGG
R: CGTCGGGAACGGGTAGAATCG |
300 |
Results
Table 2 represents the distribution of A. baumannii isolates of various types of infections. Twenty out of 150 samples (13.33%) were infected with A. baumannii. Wound infections had the highest prevalence of A. baumannii (16%), while urine had the lowest (11.66%). Statistically significant difference was seen between the type of samples and prevalence of A. baumannii (P < 0.05).
Table 2: Total distribution of A. baumannii isolates of various types of infections.
| Type of samples | No. samples collected | Prevalence of A. baumannii (%) |
| Wound | 50 | 8 (16) |
| Urine | 60 | 7 (11.66) |
| Respiratory | 40 | 5 (12.50) |
| Total | 150 | 20 (13.33) |
Figure 1 represents the results of the gel electrophoresis for detection of the virulence genes of the A. baumannii isolates of various types of infections. Table 3 shows the distribution of putative virulence genes among the A. baumannii strains of various types of infections. Results showed that bacterial strains of respiratory infections had the highest and also most variable profile of the virulence genes. Totally, csga (70%) and cnf1 (50%) were the most commonly detected virulence genes. Statistically significant difference was seen between the type of samples and prevalence of virulence genes (P < 0.05).
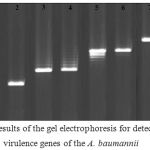 |
Figure 1: Results of the gel electrophoresis for detection of the virulence genes of the A. baumannii isolates of various types of infections. M: 100 bp ladder (Fermentas, Germany), 1, 2: Positive sample for the csgA gene and its positive control, respectively, 3, 4: Positive sample for the iutA gene and its positive control, respectively, 5, 6: Positive sample for the cnf1gene and its positive control, respectively, 7, 8: Positive sample for the cvaC gene and its positive control, respectively, and 9: Negative control. |
Table 3: Total distribution of putative virulence genes among the A. baumannii isolates of various types of infections.
| Type of samples (no positive) | Distribution of virulence genes (%) | |||
| Cnf1 | CsgA | CvaC | IutA | |
| Wound (8) | 4 (50) | 4 (50) | 1 (12.50) | 1 (12.50) |
| Urine (7) | 3 (42.85) | 5 (71.42) | – | 1 (14.28) |
| Respiratory (5) | 3 (60) | 5 (100) | 1 (20) | 3 (60) |
| Total (20) | 10 (50) | 14 (70) | 2 (10) | 5 (25) |
Table 4 indicates the pattern of antibiotic resistance of the A. baumannii isolates of various types of infections. A. baumannii strains of our study harbored the highest levels of resistance against ampicillin (100%), tetracycline (95%), gentamycin (75%) and cephalexin (60%). Prevalence of resistance against imipenem (5%) and ceftriaxone (35%) were low. Statistically significant difference was seen between the type of samples and prevalence of antibiotic resistance (P < 0.05).
Table 4: Antibiotic resistance pattern of the A. baumannii isolates of various types of infections.
| Type of samples (no positive) | Antibiotic resistance pattern (%) | ||||||||
| Lev * | Amp | Imp | Gen | Ceph | Cphx | Tet | Tr-Su | Ceft | |
| Wound (8) | 3 (37.50) | 8 (100) | 1 (12.50) | 8 (100) | 3 (37.50) | 4 (50) | 8 (100) | 4 (50) | 2 (37.50) |
| Urine (7) | 4 (57.14) | 7 (100) | – | 3 (42.85) | 3 (42.85) | 5 (71.42) | 7 (100) | 4 (57.14) | 3 (42.85) |
| Respiratory (5) | 3 (60) | 5 (100) | – | 4 (80) | 2 (40) | 3 (60) | 4 (80) | 3 (60) | 2 (40) |
| Total (20) | 10 (50) | 20 (100) | 1 (5) | 15 (75) | 8 (40) | 12 (60) | 19 (95) | 11 (55) | 7 (35) |
*Lev: levofloxacillin (5 µg/disk), Amp: ampicillin (10 u/disk), Imp: imipenem (30 u/disk), Gen: gentamycin (10 µg/disk), Ceph: cephalothin (30 µg/disk), Cplx: cephalexin (10 µg/disk), Tet: tetracycline (30 µg/disk), Tr-Su: trimethoprim/sulfamethoxazole (25 µg/disk), Ceft: ceftriaxone (30 µg/disk).
Discussion
Resistant and virulent strains of A. baumannii had a high prevalence in various types of human clinical infectious samples of immunosuppressive patients of our study. Totally, 13.33% of samples were infected with A. baumannii which was considerable. Some of the most common reasons for the high prevalence of resistant and virulent strains of A. baumannii in our study are indiscriminate and unauthorized prescription of antibiotics, daydreaming to the results obtained from the disk diffusion method, prescription of antibiotics based on the self-experience of medical practitioners, lack of proper disinfection of hospital environment, inherent nature of the bacteria that has the ability to withstand hard conditions and can survive in the surfaces and finally transmission of resistant pathogens from infected patients and workers to hospital environment and also other patients. Momtaz et al. (2015) (11) reported that A. baumannii strains were detected in 121 out of 500 human clinical samples (24.2%) which was higher than our results. Jaggi et al. (2012) (12) reported that the prevalence of A. baumannii in various types of clinical infections were 9.4% which was lower than our results. Siau et al. (1996) (13) reported that the prevalence of A. baumannii in the cases of infections in the Korean hospitals was 11% which was lower than our results. Differences in the type of samples, method of sampling, number of samples collected, method of experiment, sex and age of patients and geographical area which the samples were collected are the main factors for differences in the prevalence of A. baumannii in various investigations.
We found that bacterial strains had the high levels of resistance against ampicillin, imipenem, gentamycin, cephalexin, tetracycline and trimethoprim/sulfamethoxazole antibiotics which showed indiscriminate and unauthorized prescription of antibiotics. Management of multidrug-resistant A. baumannii infections is a countless challenge for medical practitioners and clinical microbiologists. Moradi et al. (2015) (14) showed that A. baumannii strains of human clinical infections had a high prevalence of resistance against all types of antibiotics, with the exception of carbapenems, lipopeptides, and aminoglycosides. Jaggi et al. (2012) (12) reported that the prevalence of antibiotic resistance in the A. baumannii strains of clinical samples against amikacin, gentamicin, tobramycin, aztreonem, cefipime, ceftazidime, ciprofloxacin, Levofloxacin and imepenem were 90.3%, 85.8%, 80%, 94.2%, 90.3%, 92.1%, 67.4% and 67.1%, respectively which was similar to our results. Similar findings have been reported from Denmark (15), Iran (16), Colombia (17) and China (18),
Csga, cnf1, cvaC and iutA virulence genes had a considerable prevalence among the A. baumannii strains of our clinical infections. Daryanavard and Safaei (2015) (19) reported that the total prevalence of csga, cnf1, cvaC and iutA virulence genes among the samples of UTIs were 55%, 40%, 10% and 30%, respectively which was similar to our findings. Momtaz et al. (2015) (11) reported that the prevalence of csga, cnf1, cvaC and iutA virulence genes among the A. baumannii strains of clinical infections in Iran were 12.39%, 35.53%, 21.48% and 19%, respectively which was lower than our results. Mohajeri et al. (2016) (20) showed that 40 isolates of A. baumanni strains of clinical infections had traT (80%), 17 isolates had cvaC (34%) and 8 isolates had iutA (16%) genes. These genes are the most common causes of adhesion and invasion of A. baumanni to the epithelial cells of the human organs.
Conclusions
In conclusion, we identified a large number of resistant and virulent strains of A. baumannii in the wound, urinary and respiratory infections of immunosuppressive patients hospitalized in Iranian hospitals and health care centers. Totally, respiratory infections had the highest prevalence of bacteria and also csga and cnf1 were the most commonly detected virulence genes. We found that resistance against ampicillin, tetracycline and gentamycin was maximum. Rapid diagnosis of infections caused by A. baumanni and its treatment with imipenem and ceftriaxone can reduce the risk of dissemination of A. baumanni’s infections. Judicious prescription of antibiotics according to the results of disk diffusion method can help to decrease prevalence of resistance.
References
- Wisplinghoff H, Seifert H. Epidemiology and clinical features of Acinetobacter baumannii infections in human. Berl Munch Tierarztl Wochenschr. 2014 Nov-Dec;127(11-12):447-57.
- Alsan M1,Klompas M1. Acinetobacter baumannii: An Emerging and Important Pathogen. J Clin Outcomes Manag. 2010 Aug;17(8):363-369.
- Al-Anazi KA1, Al-Jasser AM2. Infections Caused by Acinetobacter baumannii in Recipients of Hematopoietic Stem Cell Transplantation. Front Oncol. 2014 Jul 14;4:186.
- Gordon NC1, Wareham DW. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int J Antimicrob Agents. 2010 Mar;35(3):219-26.
- Lin MF1, Lan CY1. Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J Clin Cases. 2014 Dec 16;2(12):787-814.
- Eraç B1, Yılmaz FF, Hoşgör Limoncu M, Oztürk I, Aydemir S. Investigation of the virulence factors of multidrug-resistant Acinetobacter baumannii isolates. Mikrobiyol Bul. 2014 Jan;48(1):70-81.
- Eijkelkamp BA, Stroeher UH, Hassan KA, Paulsen IT, Brown MH1. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics. 2014 Nov 25;15:1020.
- Yun HC, Branstetter JG, Murray CK. Osteomyelitis in military personnel wounded in Iraq and Afghanistan. J Trauma 2008; 64(2):163-68
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. M100-S21. Wayne Pa: CLSI; 2012.
- Sambrook J, Russell D.Molecular cloning, a laboratory manual. In Cold Spring Harbor Laboratory. 3rd edition. Cold Spring Harbor, NY; 2001.
- Momtaz H, Seifati SM, Tavakol M. Determining the prevalence and detection of the most prevalent virulence genes in Acinetobacter Baumannii Isolated from hospital Infections. International Journal of Medical Laboratory 2015;2(2): 87-97.
- Jaggi N, Sissodia P, Sharma L. Acinetobacter baumannii isolates in a tertiary care hospital: Antimicrobial resistance and clinical significance. J Microbiol Infect Dis 2012, 12: 57-63.
- Siau H, Yuen KY, Wong SSY, Ho PL, Luk WK. The epidemiology of Acinetobacter infections in Hong Kong. J. Med. Microbiol. – Vol. 44 (1996), 340-347.
- Moradi J, Hashemi FB, Bahador A. Antibiotic Resistance ofAcinetobacter baumannii in Iran: A Systemic Review of the Published Literature. Osong Public Health Res Perspect. 2015 Apr; 6(2): 79–86.
- Văduva DB1,Muntean D, Lonescu G, Licker M, Văduva MB, Velimirovici D, Rădulescu M, Dumitraşcu V, Crăciunescu M, Dugăeşescu D, Horhat F, Piluţ C, Bădiţoiu L, Moldovan R. Antibiotic resistance patterns in Acinetobacter spp. strains isolated from hospital environment. Bacteriol Virusol Parazitol Epidemiol. 2008 Apr-Jun;53(2):103-7..
- Mirnejad R, Vafaei S. Antibiotic resistance patterns and the prevalence of ESBLs among strains of Acinetobacter baumannii isolated from clinical specimens. Journal of Genes, Microbes and Immunity 2013 (2013) 1-8.
- Reguero MT1,Medina OE, Hernández MA, Flórez DV, Valenzuela EM, Mantilla JR. Antibiotic resistance patterns of Acinetobacter calcoaceticus-A. baumannii complex species from Colombian hospitals. Enferm Infecc Microbiol Clin. 2013 Mar;31(3):142-6.
- Zhao S, Jiang D,Xu P, Zhang Y, Shi H, Cao H, Wu An investigation of drug-resistant Acinetobacter baumannii infections in a comprehensive hospital of East China. Ann Clin Microbiol Antimicrob. 2015; 14: 7.
- Daryanavard R, Safaei HR. Virulence genes and antimicrobial resistance properties of Acinetobacter baumannii isolated from pediatrics suffered from UTIs. Int. J. Adv. Res. Biol. Sci. 2(11): (2015): 272–279
- Mohajeri P,Sharbati S, Farahani A, Rezaei Z. Evaluate the frequency distribution of nonadhesive virulence factors in carbapenemase-producing Acinetobacter baumannii isolated from clinical samples in Kermanshah. J Nat Sci Biol Med. 2016 Jan-Jun;7(1):58-61.







