Manuscript accepted on :
Published online on: 18-12-2015
Plagiarism Check: Yes
Farshid Kafilzadeh* and Sedigheh Mokhtari
Department of Biology, Jahrom Branch, Islamic Azad University, Jahrom, Iran.
DOI : https://dx.doi.org/10.13005/bpj/402
Abstract
Phenol and phenolic compounds are the most common pollutants result of petrochemical industries. Their carcinogenic and toxic effects have been recorded on human being. Since chemical methods of removing these contaminants are expensive, applying microorganisms is suitable substitution to remove these compounds. The purpose of this research is to isolate and identify of phenol degrading bacteria from mangrove sediments in the Persian Gulf (Asaluyeh) and study of their growth kinetics and degradation level. Nine samples of mangrove sediments were collected. Isolation of bacteria was done by culture in a mineral salt-medium containing 0.5 gL-1 phenol. The growth ability of bacteria was evaluated by bacteria culture in 0.3, 0.5, 0.7, 0.9, 1, 2 and 3 gL-1 phenol concentrations. Gibbs indicator was used in order to measure the amount of phenol removal. All of isolated bacteria showed high ability in phenol removal so that Pseudomonas putida and Acinetobacter sp were able to grow in 0.9 to 2 gL-1 phenol concentrations. The findings of this research indicated that mangrove sediments have numerous phenol degrading bacteria such as; Pseudomonas putida, Acinetobacter sp, Bacillus thuringiensis, Brevibacterium iodinum, and Staphylococcus aureus.
Keywords
Phenol; Biodegradation; Pseudomonas putida; Acinetobacter sp; Mangrove sediments
Download this article as:| Copy the following to cite this article: Kafilzadeh F, Mokhtari S. Isolation and Identification of Phenol Degrading Bacteria from Mangrove Sediments in the Persian Gulf (Asaluyeh) and their Growth Kinetics Assay. Biomed Pharmacol J 2013;6(2) |
| Copy the following to cite this URL: Kafilzadeh F, Mokhtari S. Isolation and Identification of Phenol Degrading Bacteria from Mangrove Sediments in the Persian Gulf (Asaluyeh) and their Growth Kinetics Assay. Biomed Pharmacol J 2013;6(2). Available from: http://biomedpharmajournal.org/?p=2682 |
Introduction
Environmental pollutants result of industrial and agricultural activities have been one of dangerous global issues in recent years. Most of this pollutants are entered into the environment by industrial waste as a mixture of organic and inorganic contaminants and they will cause destructive effects on living systems as mutation and cancer, poisoning.
Organic contaminants result of industrial activities are phenol, polycyclic aromatic hydrocarbons, and heavy metals. These industrial activities involve petrochemical, weaving, petroleum, carbon refining, paint, and paper (1,2). One of the most dangerous ones, among these toxic contaminants is phenol which is uses as a raw material in making some kinds of chemical compounds as stains, sterilizers, and synthetic resins, and also, it is uses as indicator in chemical laboratory and an antimicrobial compound (3). Since phenol is water-soluble, wastewater result of plants which uses phenol compounds contain a significant amount of phenol which is a toxic compound for human being and the other organisms so that it is lethal in the concentrations of 5-25 mg L-1 to marine animals as fishes (2,4,5).
The UAE has one of the most stringent environmental regulations, especially those related to discharge levels. Abu Dhabi National Oil Company (ADNOC) has set a desirable limit of phenol discharge concentration of 0.01mg/l compared to that set by the EPA (USA) of 0.168mg/l (6). Therefore, it is necessary to decrease phenol concentration in industrial wastewaters to the extent that it is acceptable to environment and dangerous less by useful treatment and consumption methods (5,7,8). Phenol biodegradation has been done by applying different kinds of microbial culture in two recent decades. Many kinds of aerobic and non-aerobic bacteria are able to consume aromatic compounds as a carbon and energy resource in which phenol turns into non-toxic compounds by them in aerobic conditions. However, many of phenol treatments are as follows: biodegradation, ionic exchange, and utilization of bio-activated carbon. Also, it has been confirmed that biological degradation and bioremediation are the most useful and economical methods to remove phenol from environment and industrial wastewaters which will lead to phenol mineralization and it uses to a spectrum of phenol concentrations (7,9).
Many researches have been done in the field of phenol biodegradation in the world. A series of bacteria from several contaminated places to phenol has been isolated which are an indicator of phenol degradation activity. Some of these researches are as follows; A study and evaluation of potential of the microbial systems in order to bioremediation in Nigeria Oil Refinery wastewater by Ojumu et al. (10), isolation and identification of phenol degrading bacteria from wastewater and oil-contaminated soil by Norhani and Firdausi (11), a study of phenol biodegradation by Pseudomonas putida in polyvinyl alcohol gel in pillar bioreactor by El-Naas et al. (6), enrichment of phenol degrading moderately halophilic bacterial consortium from saline environment by Gayathri and Vasudevan (12), isolation and identification of phenol degrading bacteria from industrial wastewater and a study of their degradation level by Manafi et al. (13), isolation and identification of phenol degrading bacteria from industrial wastewaters and an evaluation of their endurance level against phenol and heavy metals synchronically by Castillo-Zacarias et al. (14), isolation, identification and characterization of elevated phenol degrading Acinetobacter sp, strain AQ5NoL1 by Ahmad et al. (15).
The purpose of this research is to isolate and identify of phenol degrading bacteria from mangrove sediments in the Persian Gulf (Asaluyeh) and study of their growth kinetics and degradation level.
Materials and Methods
Sampling
In this study the mangrove sediments in South of Iran located in Asaluyeh industrial region were examined. The sampling was conducted from surface sediments to depth 0-10 cm. The samples were put into sterile bottles, then into the containers full of ice and then transferred to lab. for enrichment in less than 24 hours (16).
Counting bacteria
Counting bacteria was done by total viable plate count method. Physiological serum with 10 -1 -10 -10 dilution was prepared by sediment samples in this method, and then, it was done surface plate in the nutrient agar contain 0.5 gL-1 phenol and nutrient agar without phenol. Also, they were incubated in 30 oC for 24-48h. Finally the number of colonies in the surface plate with and without phenol was counted (11,17).
Isolation of phenol degrading bacteria
10 g sediment sample was mixed to 100 mL of mineral-based culture containing; MnSO4.H2O, 0.01 gL-1; MgSO4, 0.1 gL-1; NaCl, 0.1gL-1; KH2PO4, 0.2 gL-1; K2HPO4, 0.4 gL-1; Phenol, 0.5 gL-1; (NH4)2SO4, 0.4gL-1; NaMoO4 .2H2O, 0.01gL-1 and incubated in 30oC with aeration for one week. Over a week, 1mL was inocubated into 100 mL new mineral-based culture (phenol broth) and incubated in 30oC for a week. Samples passage was done as until observed turbidity is as a result of bacteria activity and growth and pigment production by them as well. After the last passage, it was cultured on Agar mineral-based solid media (phenol agar), and then, monocolonies were isolated (15).
Identification of phenol degrading bacteria
According of microscopic method, the first step of identification is a study of bacteria shape and their morphology. Gram staining was used in order to determine the type of bacteria positive and negative gram. Also, biochemical tests in accordance with Bergey’s manual of systematic bacteriology were used to determine bacteria genus and specie more precisely (18).
Growth assessment of isolated bacteria in different concentrations of phenol
The best species were isolated by a study their optic absorption among phenol degrading bacteria in this method. The procedure is that, 95 mL of phenol broth media was poured into the separate erlenmeyer flasks (in phenol different concentrations). Then, 5 mL of bacterial suspension as McFarland 0.5 standard was provided and added into erlenmeyer flasks containing broth phenol media. Each bacterium is cultured in phenol different concentrations. Bacterial cultures have mineral-based solution in (0.3-0.9) gL-1 and (1-3) gL-1 phenol concentrations, and each bacterium had a control erlenmeyer flask. There are mineral salt-based cultures without phenol in the control culture. These cultures were incubated in 30oC for 7 days, and then, their optic absorption was read every 12h in frequency 600nm (19).
Evaluation of phenol elimination by isolated-bacteria
Gibbs method was used to evaluate phenol elimination by degrading-bacteria. 2,6-dichloroquinone-4-chloimide (Gibbs indicator) was used. 5 mL of surface solution of phenol broth culture was poured in a tube and by using sodium carbonate 10 %, its pH became 8. The best reaction of Gibbs indicator is done by phenol in neutral pH. Then, 25 microlitre Gibbs indicator (2,6-dichloroquinone-4-chloroimid 0.01g in 1mL pure ethanol) was added and mixed. Resultant solution was put in incubator 30 oC for 25-30 min. Following with of reracting Gibbs indicator and phenol produced blue color compound. Mineral-based solution was used to dilute samples. This solution was centrifuged (400-500 rpm for 10min), and then, its optical density was read in 630 nm. The amount of phenol removal by bacteria is achieved in accordance with standard curve of phenol. Mineral-based solution control culture contained phenol and with no bacteria (20,21,22).
Phenol standard curve
Initially solutions with phenol different concentrations were provided to establish phenol standard curve. After conducting Gibbs test, their optical absorption curve in 630 nm was drawn. Each standard curve results from 2 repetition average (22).
Statistical analysis
The results were analyzed by using the SPSS software and analysis of variance (ANOVA).
Results
The result of bacterial counting indicated that the logarithmic average of the number of bacteria in the medium containing phenol was 3.921 (cfu/g) and in the medium without phenol was 4.063 (cfu/g). The maximum number of degrading bacteria was 4.07 (cfu/g) in the station A and the minimum of them was 3.921 (cfu/g) in the station B. All of stations showed a significant difference in 1% level by Duncan test was observed in these two groups.
Isolation and identification of phenol degrading bacteria
Pseudomonas putida, Acinetobacter sp, Bacillus thuringiensis, Brevibacterium iodinum, and Staphylococcus aureus were isolated and identified in this research. Among them, the most abundance was related to Pseudomonas putida and the least of them was related to Staphylococcus aureus. The number of positive Gram bacteria was more than the negative ones. Negative gram species including Pseudomonas putida and Acinetobacter sp had too high turbidity in the culture in comparison to the other bacteria. Two above-mentioned species had more ability to degrade phenol.
A study of growth kinetics
Isolated bacteria growth in phenol different concentrations was evaluated. Bacteria growth in mineral-based culture contains different concentrations of phenol indicated that these bacteria growth in early hours of inoculation into culture is low and over 24-48h they enter into logarithmic phase, and then, optical absorption are increased by increasing turbidity result of bacteria growth, and finally, bacteria growth curve was an arising curve. Also, bacteria growth comparison in phenol different concentrations indicated that Pseudomonas putida and Acinetobacter sp growth increase as a result of increasing phenol concentration rather than phenol lower concentration. However, control culture sample without phenol showed the least turbidity in comparison with cultures with phenol (Fig 1 and 2). Growth pattern of Bacillus thuringiensis, Brevibacterium iodinum, and Staphylococcus aureus were the same. Their growth was well by phenol concentration of 0.9 gL-1 (Fig. 3, 4 and 5). High turbidity and growth was observed as a result of all of 5 bacteria inoculation into culture in 1-3 gL-1 phenol concentrations (Fig. 6).
An evaluation of phenol removal and a comparison of its removal rate by bacteria
The findings indicated that phenol was decreased in half by bacteria during 48h so that over 48h Pseudomonas 83%, Acinetobacter 62.8%, Staphylococcus aureus 62%, Brevibacterium iodinum and Bacillus thuringiensis 52%, and bacterial mixture 37.8% degraded phenol added into culture. The most effectiveness observed in Pseudomonas putida and Acinetobacter sp. Similarly, phenol was removed 100% over 108 and 120h. Another bacteria degraded phenol 94% (Brevibacterium iodinum), 93% (Staphylococcus aureus), 91% (Bacillus thuringiensis) for 7 days. The least of removal level related to all 5 bacteria culture in which the level of phenol removal was 81.2% (Fig. 7 and 8).
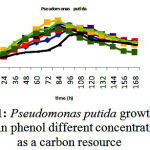 |
Figure 1: Pseudomonas putida growth curve in phenol different concentrations as a carbon resource
|
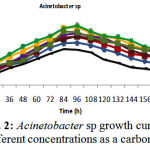 |
Figure 2: Acinetobacter sp growth curve in phenol different concentrations as a carbon resource
|
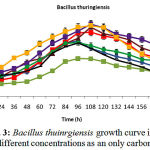 |
Figure 3: Bacillus thuinrgiensis growth curve in phenol different concentrations as an only carbon resource
|
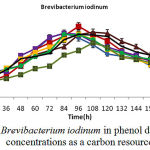 |
Figure 4: Brevibacterium iodinum in phenol different concentrations as a carbon resource
|
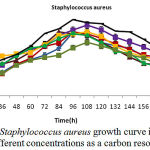 |
Figure 5: Staphylococcus aureus growth curve in phenol different concentrations as a carbon resource
|
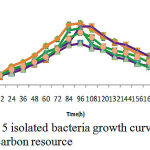 |
Figure 6: A mixture of 5 isolated bacteria growth curve in phenol different concentrations as a carbon resource
|
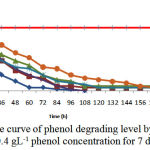 |
Figure 7: The curve of phenol degrading level by bacteria in 0.4 gL-1 phenol concentration for 7 days
|
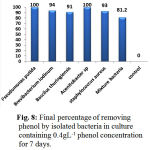 |
Figure 8: Final percentage of removing phenol by isolated bacteria in culture containing 0.4gL-1 phenol concentration for 7 days.
|
Discussion
In a research by Gayathri et al. the bacterial consortium was isolated from mixtures of soil from phenol contaminated sites and as well as from areas having proximity to saline environment. The 16S r-RNA gene analysis and biochemical tests showed that the bacterial consortium contained six bacterial strains, which were identified as Bacillus cereus, Arthrobacter sp., Bacillus licheniformis, Halomonas salina, Bacillus pumilus and Pseudomonas aeruginosa (12).
Phenol degrading bacteria were isolated of Siberia soils by Koutny et al. (19). They concluded that a dominant species in phenol degradation is Pseudomonas particularly putida in these soils. Also, their distribiution in the soil and phenol or phenolic compounds degradation power were confirmed by numerous researchers as Williams and Sayers (23) and Powoloski and Shinger (24). In the present research, the best and the effectiveness phenol degrading bacteria are as follows; Pseudomonas putida, Brevibacterium iodinum, Bacillus thuringiensis, Acinetobacter sp, and Staphylococcus aureus that all of them adjust with the result of other researches except two latter species. Klebsiella pnemoniae, Psudomonas aeruginosa, Escherichia coli were isolated from contaminated industrial effluents by Castillo-Zacarias et al. (14). These bacteria showed high endurance against 1000 mg L-1 phenol concentration. RWC–sma and RWC–crl were isolated from wastewater and ISC–Ycr and ISC–Tra isolated from contaminated soil to oil by Norhani and Firdausi (11). According to Bergey’s manual of systematic bacteriology book, there is a 86 % probability in which isolated species belong to Pseudomonas, Alcaligenes, and Acinetobacter. Also, degrading bacteria were experimented sequentially from phenol different concentrations to 1000 mg L-1. Putting bacteria against phenol increasingly concentrations are used to isolations adjustment. Thus, the best phenol degrading cases are more compatible bacteria and the ones which are able to phenol degrading in higher concentrations. In the current research the growth of Pseudomonas putida, Acinetobacter sp, and Bacillus thuringiensis will be increased by increasing phenol concentration, and then, its toxic effects appeared in concentrations above 2000 mgL-1.
Oil-consuming Acinetobacter AQ5N0L1 was isolated of contaminated soil to oil so that this bacterium could degrade phenol in 1500 mgL-1 concentration (15). Phenol removal of industrial wastewater in the existence of Alcaligene faecalis was studied by Manafi et al., and then, they concluded that phenol concentration of 1200 mgL-1 had an inhibitory effect on cell growth and phenol degradation (13). In the current research Acinetobacter sp was able to grow in phenol culture by concentration of 2000 mgL-1. Also, growth inhibitory effects appeared in higher concentrations.
Biodegradation by Pseudomonas Putida established in alcohol polyvinyl gel was done in a pillar bioreactor by El-Nass et al. (6). This bacterium was able to consume aromatic compounds as a resource of carbon and energy. Experimental findings indicated that temperature, phenol initial concentration, and biomass abundance will affect the ability of this bacterium to degradation. The level of biodegradation has been optimized in 30oC and concentration phenol of 75 mgL-1. Higher phenol concentrations controlled biomass, and also, it decreased phenol biodegradation. Selected concentration and result of this research is inconsistent with the present research. Different methods have been applied to measure the amount of phenol removal. For example the amount of bacterial phenol removal was estimated by 4-amino antipirin and in accordance with colorimetric method by Wantanabe et al. (25). Also, this method was used to measure the amount of rsidual phenol by Ojuma et al. (10) and Ahmad et al. (15). HPLC method was applied to measure the bacterial degradation of phenol by Selvaratnam et al (26) and Mohite et al. (27).
Gas chromatography method was applied to measure the amount of phenol removal of bacteria by Antizar-Ladislao and Galil (28). Colorimetric method in accordance with Gibbs indicator was applied to measure the amount of phenol degradation by isolated bacteria from Parishan Lake by Kafilzadeh et al. (29). Gibbs method by using 2,6-dicholoroquinone-4-chloroimide was used to measure phenol biodegradation in the present research. In a research by Manafi et al. Alcaligenes faecalis was able to remove phenol by 1000 mg L-1 concentration so that phenol was degraded by bacteria 79% in 400 mg L-1 and 92 % in 530 mg L-1 of phenol concentration (13).
In a research by Castillo-Zacarias et al. Klebsiella pneumoniae, Pseudomonas aeruginosa, and E.coli showed high endurance against phenol concentration of 1000 mg L-1 (the percentage of removal was 23%-78% for 24h) (14).
Phenol concentrations of 300 mgL-1 and 200 mgL-1 were applied to isolate phenol degrading bacteria respectively by Wantanabe et al. (25) and Whitely et al. (30). In the current study the most amount of phenol degradation was reported in 600 mgL-1 and 500 mgL-1 phenol concentrations. In this research 500 mgL-1 initial concentration was applied to isolate phenol degrading bacteria. The growth was increased by increasing concentration, and also, concentrations higher than 500 mgL-1 caused bacteria growth stimulation and degradation. It was consistent with Manafi et al. (13) and inconsistent with Whitely et al. (30) findings.
Bacillus Stearothermophilus which is able to degrade endurance was isolated by Lee et al. (31). Also, a species of Bacillus is able to grow by phenol concentration 1000 mgL-1 in the present research.
Phenol bioremediation in bioreactor was studied by Singh and Fulekar (32). The results of this research indicated that phenol concentration more than 1000 mgL-1 was removed 100% for 7 days. Also, phenol concentration of 500 mgL-1 was removed 100% during 120h. In the current research phenol concentration of 400 mgL-1 was eliminated by Pseudomonas putida and Acinetobacter sp 100% during 96 and 108h.
Conclusion
Mongrove sediments in the Persian Gulf are a suitable bed in order to isolate phenol degrading bacteria. Pseudomonas putida and Acinetobacter sp isolated of these sediments were the most powerful ones in phenol degradation. Therefore, bioremediation by them is the cheapest and the most economical method to eliminate these contaminants and a suitable substitution to expensive physicochemical methods.
References
- Keith L.H., Telliand W.A. Priority pollutants. Environ. Sci. Technol., 1979; 13: 416-423.
- Collins L.D. and Daugulis A.j. Characterization and optimization of a two-phase partitioning bioreactor for the biodegradation of phenol. Appl. Micobiol. Biotechnol., 1997; 48(1): 18-23.
- Kumar A., Kumar S. and Kumar S. Biodegradation kinetics of phenol and catechol using Pseudomonas putida 1194. Biochem. Eng. J., 2005; 22: 151-159.
- Ghadhi S.C., Sangodkar U.M.X. Potentials of pseudomonas cepacia PAA in bioremediation of aquatic wastes containg phenol. Proc. Natl. Symp. Frontiers in App. Environ. Microbiol., Dec. Chochin, 1995; 11-13.
- Prieto M.B., Hildalgo A., Serra J.L. and Llama M.J. Biodegradation of phenol by Rhodococcus erythropolis UPV-1 immobilized on Biolite. J. Biotechnol., 2002; 97(1): 1-11.
- El-Nass M.H., Al-Muhtaseb S.A., Makhlouf S. Biodegradation of phenol Pseudomonas putida immobilized in poly vinyl alcohol (PVA) gel. J. Hazard. Mater., 2009; 164(2-3):720-725.
- Hannaford A.M. and Kuek C. Aerobic batch degradation of phenol using immobilized Pseudomonas putida. J. Indust. Microbiol. Biotechnol., 1999; 22:121-126.
- Kowalska M., Bodzek M. and Bohdziewicz J. Biodegradation of phenols and cyanides using membranes with immobilized microorganisms. Process. Biochem., 1998; 33(2):189-197.
- Nuhoglu A. and Yalcin B. (2004) Modeling of phenol removal in a batch reactor. Process. Biochem., 2008; 40: 233-239.
- Ojumu T.V., Bello O., Sonibare J.A. and Solomon B. Evaluation of microbial systems for bioremediation of petroleum refinery effluents in Nigeria. Afr. J. Biotechnol., 2005; 4(1): 31-35.
- Norhani J. and Firdausi R. Microbial consortia from residential wastewater for bioremediation of phenol in a chemostat. J. Technol., 48(F): 51-60.
- Gayathri K.V., Vasudevan N. Enrichment of phenol degrading moderately halophilic bacterial consortium from saline environment. J. Bioremed. Biodegrad., 2010; 1:104.
- Manafi M., Mehrnia M.R., Sarrafzadeh M.H. Phenol removal from synthetic wastewater by Alcaligenes faecalis. International Journal of Chemical and Environmental Engineering., 2011; 2(2): 103-107.
- Castillo-Zacarias C.J., Suarez-Herrera M.A., Garza-Gonzalez M.T., Sanchez-Gonzalez M.N. And Lopez-Chuken U.J. Biosorption of metals by phenol -resistant bacteria isolated from contaminated industrial effluents. Afr. J. Microbiol. Res., 2011; 5(18): 2627-2631.
- Ahmad S.A., Mohd Arif S., Noorliza M., Mohd Yunus A.S., Nor Aripin S. Isolation, identification and characterization of elevated phenol degrading Acinetobacter sp, strain AQ5NoL1. Aust. J. Basic & Appl. Sci., 2011; 5(8): 1035-1045.
- Mashreghi M., Mashreghi K. Characterization of bacteria degrading petroleum derivatives isolated from contaminated soil and water. J. Sci. Islam. Repub. Iran., 2005; 16(4): 317-320.
- Udeaani T.K.C., Obroh A.A., Okwuosa C.N., Achukwu P.U., Azubike N. Isolation of bacteria from mechanism work shop soil environment contaminated with used engine oil. Afr. J. Biotechnol., 2009; 8(22): 6301-6302.
- Garrity G.M., Brenner D.J., Krieg NR., Staley J.T. Bergey’s manual of systematic bacteriology. 2nd ed. New York: Spriger: 2005; P. 323-840.
- Koutny M., Ruzicka J., Chlachula J. Screening for phenol-degrading bacteria in the Pristine soils of south Siberia. Appl. Soil Ecol., 2003; 23(1): 79-83.
- Quintana M.G., Didion C., Dalton H. Colorimetric method for a rapid detection of oxygenated aromatic biotransformation product. Biotechnol. Tech., 1997; 11(8): 585-587.
- Ali S., Fernandez-Lafuente R.A. and Cowan D.A. Meta-pathway degradation of phenolics by thermophilic Bacilli. Enzyme Microb. Technol., 1998; 23(7-8): 462-468.
- Rhee S.K., Chang J.H., Chang Y.K., Chang H.N. Desulfurization of DBT and Diesel Oils by a newly isolated Gordona Strain CYKS1. Appl. Environ. Microbiol., 1998; 64(6): 2327-2331.
- Williams PA., and Sayers JR. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation., 1994; 5(3-4): 195-217.
- Powlowski J., and. Shingler V. Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation., 1994; 5(3-4): 219-236.
- Wantanabe K., Hino S. and Takahashi N. Effect of exogenous phenol-degrading bacteria on performance and ecosystem of activated sludge. J. Ferment. Bioeng., 1996; 82(31):291-298.
- Selvaratnam S., Schoedel B.A., Mcfarland B.L., Kulpa C.F. Application of revers transcriptase PCR for monitoring expression of the catabolic dmp N gone in phenol-degrading sequencing batch reactor. Appl Environ Microbiol., 1995; 61(11): 3981-3985.
- Mohite B.V., Jalgaonwala R.E, PAWAR S., Morankar A. Isolation and characterization of phenol degrading bacteria from oil contaminated soil. Innovat. Rom. Food Biotechnol., 2010; 7: 61-65.
- Antizar-Ladislao B., Galil. N.L. Enhanced in situ bioremediation of phenol in bioestimulated saturated and unsaturated sand-bed columns. Water Environ. Res., 2006; 78(13): 2447-2455.
- Kafilzadeh F., Farhangdoost M.S., Tahery Y. Isolation and identification of phenol degrading bacteria from lake Parishan and the growth kinetic assay. Afr. J. Biotechnol., 2010; 9(40): 6721-6726.
- Whitely AS.,Wiles S.,Lilley AK., Philp J., Bailey MJ. Ecological and physiological analyses of Pseudomonad species within a phenol remediation system. J Microbiol Methods., 2001; 44(1): 79-88.
- Lee D-H., Noh S-A. and Kim C-K. Development of molecular biological methods to analyze bacterial species diversity in freshwater and soil ecosystem. J. Microbiol., 2000; 38(1):11-17.
- Singh D. and. Fulekar M.H. Bioremediation of phenol using microbial consortium in bioreactor. Innovat. Rom. Food Biotechnol., 2007; 1: 31-36.







