Manuscript accepted on :20 May 2010
Published online on: 24-11-2015
Plagiarism Check: Yes
V. Padmapriya* and N. Anand
Centre for Advanced Studies in Botany, University of Madras, Guindy Campus, Chennai - 600 025 (India).
Abstract
Cytosolic iron-containing superoxide dismutase (FeSOD; EC: 1.15.1.1) was purified to homogeneity from the cyanobacterium, Anabaena variabilis Kutz. The stepwise purification consisted of ammonium sulphate precipitation, anion-exchange chromatography and gel-filtration column chromatography. The purified enzyme was hydrogen peroxide-sensitive, confirming the enzyme to be of the iron type. Purification of 269 folds was achieved and specific activity of 700 U/mg protein obtained. Homogeneity of the purified enzyme by denaturing electrophoretic analysis showed the presence of a single band in the range of 41.4 KDa. Metal analysis of the purified enzyme showed the presence of 0.88 g atoms of iron in 41400 g of the purified enzyme. The purified SOD enzyme from A. variabilis had pH optimum of 7.4 and was active even after 1 h incubation at pH 7-7.4. The enzyme was heat-labile and was susceptible to dimethyl sulfoxide (DMSO) treatments above 20%. The UV-visible spectrum of the purified enzyme at room temperature had peaks at 253 nm and 267 nm.
Keywords
Purification; characterization; iron superoxide dismutase; cyanobacteria; Anabaena variabilis
Download this article as:| Copy the following to cite this article: Padmapriya V, Anand N. Purification and Characterization of Iron-Containing Superoxide Dismutase from Anabaena Variabilis Kutz ex Born. et Flah. Biomed Pharmacol J 2010;3(2) |
| Copy the following to cite this URL: Padmapriya V, Anand N. Purification and Characterization of Iron-Containing Superoxide Dismutase from Anabaena Variabilis Kutz ex Born. et Flah. Biomed Pharmacol J 2010;3(2). Available from: http://biomedpharmajournal.org/?p=1571 |
Introduction
Cyanobacteria possess extraordinary metabolic processes and mechanisms to survive which has helped them tide over the geological upheavals from the Precambrian era to the present day with success. They withstand extremes of temperature, desiccation, illumination, radiations, salinity, pH, nutrient constraints and toxicants, occur in diverse habitats and have evolved alternate metabolic pathways and suitable regulatory mechanisms to tide over extreme conditions. Intracellular reactive oxygen species (ROS) rises due to stress resulting in oxidative damages and the redox enzyme-superoxide dismutase (SOD), scavenges the undesirable oxygen radicals by converting them into peroxides thereby avoiding the formation of the highly toxic hydroxyl radicals (1). The active metal site of the enzyme shifts between the oxidized and reduced state and enables the enzyme to act as an oxidant or reductant. This ping-pong mechanism of the enzyme activity thus accounts for the near diffusion-limited rates of approximately 109M-1s-1(2). Cyanobacteria have a highly-evolved and well-regulated antioxidant enzyme cascade consisting of SODs, catalases and peroxidases for the removal of ROS. SODs are highly valued as therapeutic enzyme drugs (3, 4) and were first commissioned in 1985 to be researched as an enzyme drug in the USA for defence of donor organs against oxidative stress during periods of ischemia and reperfusion (5). In this study, iron-containing SOD was purified from the heterocystous, nitrogen-fixing, filamentous cyanobacterium, Anabaena variabilis Kutz ex Born. et Flah and characterized.
Materials and Methods
Growth conditions
Cultures were grown and maintained in BG11 medium (6), at a temperature of 27°C±1°C under fluorescent illumination of 30-40 µEm-2s-1 provided by fluorescent tubes (Philips Trulite, Col 82). Cultures were exposed to a 12 h light / 12h dark cycle photoperiod controlled by automatic timers (Sangmo Weston Ltd, S650 313F model) and were swirled manually for five minutes, thrice daily. The cultures were estimated for protein (7) and SOD (8). The SOD activity was expressed in units (U) where 1U corresponded to the amount causing half the maximum inhibition of nitroblue tetrazolium (NBT) to blue formazan. The activity was calculated using the formula- U/mL = [(V0/v) – 1] (dilution factor); V0: A560 of control, v:A560 of sample. The results were expressed as U mg protein-1. The polyacrylamide gel electrophoresis (PAGE) for native gel-assay was carried out under a constant current of 10 mA under non-denaturing conditions. SOD bands appear colorless against a blue background and isozymes were detected by the method of Beyer and Fridovich (9). The gels were cut and soaked in 2 mM of KCN or 2 mM of H2O2 for 10 mins prior to soaking in NBT. FeSOD is inactivated by H2O2, Cu-ZnSOD is inactivated by KCN while MnSOD remains unaffected by these compounds. Sodium dodecyl sulphate (SDS) PAGE was also carried out for determination of the molecular weight of the purified protein (10). All the experiments were conducted in triplicates. Standard deviation and One-Way ANOVA were used for the statistical analysis of the data.
Purification of SOD
Cells were harvested by centrifugation at 3024 g for 10 minutes in a Remi Research centrifuge. They were washed twice in Buffer A [Tris HCl, pH 7.8, 20mM; NaCl 10mM] and suspended in a minimum volume of the Buffer A containing 100μM of Phenyl methane sulfonyl flouride (PMSF) prior to lysing in a sonicator. The homogenate was centrifuged at 12000 g for 1h at 4ºC in a Beckman L7-65 Ultracentrifuge. To the clear supernatant, solid ammonium sulphate was added to a saturation of 60%. The solution was stirred and kept in ice for 30 minutes. Ultra centrifugation at 27216 g for 1h at 4ºC was carried out again and the pellet containing the phycobilin pigments was discarded. The supernatant was brought to a final saturation of 80% with solid ammonium sulphate, the solution was stirred, kept at 4ºC for 30 minutes to 1h and then centrifuged at 40000 g. The pellet containing the enzyme activity was resuspended in buffer A and dialyzed against the same buffer overnight at 4ºC. To remove the precipitant formed during dialysis, the sample was centrifuged at 3000 g for 15 minutes at 4ºC. The supernatant containing the partially purified enzyme was taken for further purification. DEAE cellulose was used as the matrix for the first chromatographic separation to separate MnSOD from FeSOD based on their different pI values. The cellulose was activated by washing with 0.1N NaCl and 0.1N HCl. The cellulose was finally given an equilibrating buffer wash with Buffer A and the slurry was poured into the column of dimensions 13cm x 1.4 cm. The column was left undisturbed so that the cellulose settled under gravity. The column was again washed with buffer (~ 2 column volumes) and the pH of the wash-out was checked to ascertain the equilibrium of the ion exchange material. The concentrated enzyme sample obtained from the initial fractionation step was loaded on to the column and was washed with 3 column volumes of buffer solution to remove the unbound proteins. The enzyme was then eluted with a linear NaCl gradient from 0.01M to 0.5M in the buffer. Fractions of 2 mL each were collected. The eluted fractions were assayed for enzyme activity along with protein content. The fractions containing enzyme activity were pooled, dialyzed against Buffer A and concentrated by freeze-drying. They were then stored at 4ºC till further use. Sephadex G75 gel matrix was used for further purification. The dry gel powder was swollen with 3 volumes of the equilibrating buffer and poured in to the column (21 cm x 1.4 cm). The flow rate of the column was 0.8 mL/min and fractions of 0.5 mL each were collected. The enzyme sample obtained from the previous chromatographic fractions was loaded onto the column and washed with Buffer A. The fractions were analyzed for protein and enzyme. The active fractions were pooled, dialyzed and concentrated using Centricon YM-30 filter device. The retentate and the filtrate were checked for enzyme activity and used in experiments for characterization of the purified enzyme.
Characterization of sod
For the characterization studies, 1 U of the purified enzyme was taken to evaluated the optimum pH, stability of the enzyme in varying pH and temperatures and internal stabilization of the enzyme. For the cross reactivity studies with antibodies and other inhibitors, I U of the purified enzyme was incubated with 1mM H2O2, 1mM KCN and 20μg MnSOD antibodies for 10 mins at 4ºC. The UV-Visible spectrum of the purified enzyme was analyzed in a UV5704SS Double Beam Spectrophotometer (Electronic Corporation of India). Metal analysis of the purified enzyme was carried out in an Atomic Absorption Spectrophotometer (Perkin-Elmer AA700 Spectrometer) for the detection of iron and manganese elements. The purified enzyme sample was mixed with twice the volume of tri-acid mixture (conc. H2SO4: HNO3: HClO4 mixed in the ratio 2:9:1) and the total volume was made up to 10mL using sterile water and left undisturbed overnight. The sample was then centrifuged at 12096 g for 5 mins and the clear supernatant taken up for metal analyses.
Reagents
The matrix for gel filtration chromatography and the authentic enzyme sample were purchased from Sigma-Aldrich, India. Protein molecular weight markers were obtained from Bangalore Genei India Limited. All other chemicals used were of the high analytical grade obtained from Merck India Limited, Loba Chemie, Qualigens and SRL India Limited. Centricon ultra filters, 0.22μM membrane filters, PD-10 desalting columns were a kind gift from Dr. Vijaya Velu.
Results
Purification
The summary of the purification of SOD from A. variabilis is presented in Table 1. When passed through the ion exchange column (DEAE cellular matrix-anion exchanger), purification of 15 folds was achieved, with the enzyme showing a specific activity of 39 U/mg protein. Although 4 major peaks of enzyme activity were observed (Fig.1) only fractions 37-45 were taken up for the next study. The concentrated sample was then passed through the gel filtration column with Sephadex G-75 as the gel matrix (Fig.2). The enzyme was detected in fractions 28-39. The fractions were pooled and concentrated using centrifugal filter device (Centricon YM-30). Enzyme activity was present in the retentate fraction. The specific activity of the enzyme increased to 700 U/mg protein. A purification fold of 269 was obtained and the yield was 4.5%. This may be due to the very low amount of soluble protein extract in the first step.
Table 1. Purification of hydrogen peroxide-sensitive FeSOD from A. variabilis.
| Sl.No. | Fractions | Total protein (mg) | Total enzyme units (U) | Specific activity (U/mg) | Purification fold | Yield (%) |
| 1 | Soluble extract | 120 | 312 | 2.6 | 1 | 100 |
| 2 | 60% (NH4)2SO4 supernatant | 40 | 280 | 7.0 | 2.7 | 89.7 |
| 3 | 80% (NH4)2SO4 pellet | 10.5 | 265 | 25.2 | 9.7 | 85 |
| 4 | IE chromatography | 0.78 | 30.4 | 39 | 15 | 9.7 |
| 5 | Gel filtration | 0.02 | 14 | 700 | 269.2 | 4.5 |
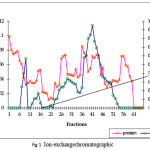 |
Figure 1: Ion-exchange chromatographic separation of FeSOD from A. variabilis using DEAE cellulose matrix and NaCl gradient for elution.
|
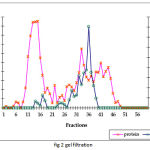 |
Figure 2: Gel filtration chromatographic separation using Sephadex G-75 gel matrix for fractions obtained from IE chromatograph.
|
Table 2: Metal analysis of the purified FeSOD.
| Element | Ppm | |
| 1. | Manganese (blank) | 0.014 |
| 2. | Manganese (enzyme) | 0.164 |
| 3. | Iron (blank) | BDL* |
| 4. | Iron (enzyme) | 9.838 |
BDL*: Below detectable levels
Characterization
pH optima, determined by assaying 1 U of the purified enzyme in reaction mixture with varying pH revealed that optimum results were obtained in pH 7.4 where SOD showed nearly 68% inhibition of formazan formation. (Fig.3). Stability of SOD as a function of pH showed that there was total absence of enzyme activity in pH 7.8 and 8.2. The enzyme was stable at pH 7.0 and 7.4 showing nearly 50-60% inhibition in the reduction of the tetrazolium salt (Fig.4). Stability of SOD as a function of temperature revealed that at pH 7.8 and a 1 h incubation, the enzyme was stable at 37ºC with 50% inhibition of formazan formation and at 45ºC with 46% inhibition of formazan formation. The enzyme was totally deactivated at 60ºC. Low enzyme activities were observed at 30ºC (27% inhibition) and 52ºC (15% inhibition) (Fig.5). Stability of SOD towards dimethyl sulphoxide (DMSO) was evaluated to study the internal stabilization of the enzyme. 1U of the purified enzyme incubated in varying concentrations of DMSO for 1 h on ice at pH 7.8 revealed that the enzyme activity was unaffected by 20% DMSO but was reduced greatly at 40% DMSO (Fig 6). At higher concentrations, the enzyme was totally inactivated. Stability of SOD towards inhibitors and antibodies revealed that the enzyme activity was unaffected by the presence of anti-MnSOD or KCN but was totally inactivated by 1mM of H2O2, suggesting a probability of an iron-containing protein (Fig. 7). SDS-PAGE analysis of the purified enzyme revealed a single major band at 41.4 KDa which coincided with one of the 4 bands seen in the standard FeSOD (Plate 1). The presence of the purified enzyme, crude enzyme and the standard FeSOD electrophoresed simultaneously on non-denaturing 10% resolving gel and subjected to the inhibitors-1mM KCN and 1mM H2O2 are also shown in Plate 1. In the presence of KCN, the enzyme bands in purified and crude samples are clearly distinguished, which is not the case in 1mM H2O2-treated gel strip. In the latter, only the top band is visible in the crude enzyme sample indicating the presence of MnSOD. Metal analyses of the purified enzyme revealed the presence of Fe at 9.38 ppm and manganese at 0.16 ppm (Table 2). The UV-visible spectrum of the purified enzyme revealed very low absorption at 280 nm, probably due to a very low content of tyrosine and tryptophan residues. Auto peaks were observed at 253 nm and 265 nm (Plate 2). The UV-Visible spectrum of the standard FeSOD is also presented.
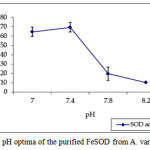 |
Figure 3: pH optima of the purified FeSOD from A. variabilis.
|
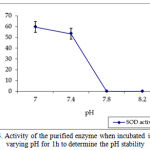 |
Figure 4: Activity of the purified enzyme when incubated in varying pH for 1h to determine the pH stability.
|
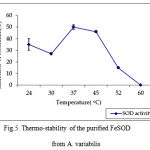 |
Figure 5. Thermo-stability of the purified FeSOD from A. variabilis.
|
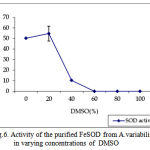 |
Figure 6: Activity of the purified FeSOD from A.variabilis in varying concentrations of DMSO
|
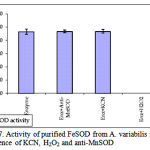 |
Figure 7: Activity of purified FeSOD from A. variabilis in the presence of KCN, H2O2 and anti-MnSOD
|
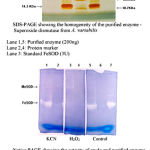 |
Plate 1:
|
 |
Plate 2:
|
Discussion
The first report of the isolation and purification of SOD was given by Lumsden and Hall (11). The SOD was purified from Spiruina sp., was of the iron type and the specific activity was 6 U/mg protein. Misra and Keele Jr. (12) reported the purification of FeSOD from Plectonema boryanum with a molecular weight around 36,500 and specific activity of 2900 U/mg protein. Simultaneously, another group of scientists (13) reported the presence of MnSOD and FeSOD enzymes from P. boryanum. The molecular weight was 41.7 KDa and the enzyme showed an absence of Fe-S clusters. FeSOD was shown to be composed of 2 subunits of equal size without a disulphide bridge between the subunits. Lumsden et al., (14) purified and studied the physicochemical properties of the FeSOD from Spirulina platensis. The stability of the enzyme was studied as a function of pH, temperature and towards DMSO and hydrogen peroxide. The enzyme was functional at temperatures of 70ºC and plots of log (activity) against time at 70ºC gave straight lines showing half lives of 6-5 minutes for Spirulina FeSOD enzyme. Cseke et al., (15) reported the isolation and purification of a FeSOD from Anacystis nidulans with a molecular weight of 37.5 KDa, consisting of 2 subunits of equal molecular weight. EPR and crystal studies showed that the symmetry of the Fe3+ ion in the SOD molecule was nearly rhombic. FeSOD was also isolated from A. cylindrica (16) and the molecular weight was nearly 40 KDa.
In the present study, the purification and characterization of SOD in A. variabilis was carried out. The purified enzyme was sensitive to hydrogen peroxide and was confirmed to possess iron in the prosthetic group. Purification of 269 folds was achieved and specific activity of 700 U/mg protein was obtained. Homogeneity of the purified enzyme showed the presence of a single band in the range of 41.4 KDa. Interestingly, immunoblot studies in Chroococcidiopsis for FeSOD showed the presence of a band of 38 KDa instead of the usual 20-22 KDa, which is the molecular weight normally attributed to FeSOD subunits (17). Canini et al., (18) also showed the presence of the FeSOD band detected by immunoblot method at nearly 38 KDa in the early degenerate cells of Microcystis aeruginosa. The result in our study was reproducible and corresponded to one of the 3 bands present in the authentic sample.
The UV-visible spectrum of the purified enzyme at room temperature did not show any peak at 280 nm, which may be due to the very low presence of aromatic amino acids. Peaks at 253 nm and 267 nm were seen. Spectral studies on the FeSOD of Plectonema boryanum at room temperature showed peak at 280 nm, shoulders at 260 nm and 290 nm and weak absorption from 320 nm-600 nm with a shoulder at 350 nm (13). In the present study, no absorption was observed above 300nm. In the earlier reports on bovine hemocuprein absence of a peak around 280 nm indicated almost zero levels of tryptophan and tyrosine residues (19). In the present study, the atomic absorption analysis for metal content of the purified enzyme showed 0.88g atoms of Fe per 41400g of the pure enzyme with no detectable level of Mn. This lower value of iron may possibly be due to loss of the element during isolation. Atomic absorption spectrometer results on the purified FeSOD enzyme from P. boryanum indicated 0.94 mole of Fe per 36500 g of the enzyme (12) whereas metal analysis indicated the occurrence of 2g atoms of Fe per mole of the enzyme in the experiments carried out by Asada et al., (13) in P. boryanum. Cyanobacteria are able to avail even tightly bound iron but in the presence of very strong chelators, the iron may become biologically unavailable resulting in an increase in the stress leading to an increase in the antioxidant enzymes (20).
In this study, the purified SOD enzyme from A. variabilis, which had pH optimum of 7.4, was active even after incubation for 1hr at pH 7.0 and 7.4. The enzyme was not active at 60ºC proving to be heat-labile and was susceptible to DMSO treatments above 20% since DMSO is considered to be effective in disrupting hydrophobic forces. Lumsden et al., (14) reported that the FeSOD from Spirulina platensis was rapidly inactivated at pH above 10.8 when incubated for 30 mins and the enzyme was very unstable below pH 5.0 (determined using the cytochrome C reduction assay). The Spirulina enzyme was stable in up to 55% DMSO (v/v). Forman and Fridovich (21) when studying the rate constants of the SOD enzymes observed reduction in the activity of the FeSOD enzymes of E.coli at increasing pH and the stability of the enzymes subjected to a period of 5-10 minutes exposure in the pH range 6 to 10.2 showed no loss of activity.
The present study paves the way for procuring monoclonal antibodies against the purified FeSOD for further use in immunological studies and in molecular biological studies.
Acknowledgements
PV was a recipient of the CSIR-Senior Research Fellowship from the Govt. of India during the tenure of this study. This work forms a part of the doctoral thesis submitted by PV to the University of Madras, Chennai, India.
References
- Wolfe-Simon, F., D. Grzebyk and O. Schofield. 2005: The role and evolution of superoxide dismutases in algae. J. Phycol. 41: 453-465.
- Falconi, M., P. O’Neill, M. E. Stroppolo and A. Desideri. 2002. Superoxide dismutase kinetics. Enzymol. 349: 38-49.
- Bannister, J.V., Bannister, W.H., Rotilio, G. Aspects of the structure, function and applications of superoxide dismutase. CRC Critical Rev. Biochem. 1987; 22: 111-180.
- Proctor, P.H., Reynolds, E.S. Free radicals and disease in man. Physiol. Chem. Phy. Med. NMR, 1984; 16: 175-195.
- Vellard, M. The enzyme as drug: Application of enzymes as pharmaceuticals. Curr. Opinion in Biotech. 2003; 14:1–7.
- Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111: 1-61.
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248-254.
- Beauchamp, C. and I. Fridovich.1971. Improved assays and assay applicable to acrylamide gels. Anal. Biochem.. 44: 276-287.
- Beyer Jr., W. F. and I. Fridovich. 1987. Assaying for superoxide dismutase activity: some large consequences of minor changes in conditions. Analytical Biochemistry. 161: 559-566.
- Laemmile, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London). 227: 680-685.
- Lumsden, J. and D. O. Hall. 1974. Soluble and Membrane bound superoxide dismutases in a blue-green alga (Spirulina) and spinach. Biochem. Biophy. Res. Comm. 58: 35-49.
- Misra, H. P. and B. B. Keele Jr. 1975. The purification and properties of superoxide dismutase from a blue –green alga. Biochimica et Biophysica Acta. 379: 418-425.
- Asada, K., K. Yoshikawa, M. Takahashi, Y. Maeda and K. Enmanji. 1975. Superoxide dismutase from a blue-green alga, Plectonema boryanum. J. Biol. Chem. 250: 2801-2807.
- Lumsden, J., R. Cammack and D. O. Hall. 1976. Purification and physiochemical properties of superoxide dismutase from two photosynthetic microorganisms. Biochemica et Biophysica Acta. 438: 380-392.
- Cseke, C., L. L. Horvath, P. Simon, G. Borbely, L. Keszthelyi and G. L. Farkas. 1979. An iron-containing superoxide dismutase from Anacystis nidulans. Biochem. 85: 1397-404.
- Canini, A., P. Civitareale, S. Marini, M. G. Caiola, and G. Rotilio. 1992b. Purification of iron superoxide dismutase from the cyanobacterium Anabaena cylindrica and localization of the enzyme in heterocysts by immuno gold labeling. Planta. 187: 438-444.
- Caiola M. G., A. Canini and R. Ocampo-Friedmann. Iron Superoxide dismutase (Fe-SOD) localization in Chroococcidiopsis sp. (Chroococcales, Cyanobacteria). Phycologia 35: 90-94.
- Canini, A., D. Leonardi and M. G. Caiola. 2001. Superoxide dismutase activity in the cyanobacterium Microcystis aeruginosa after surface bloom formation. New Phytol. 152: 107-116.
- Mc Cord J. .M. and I. Fridovich. 1969. Superoxide dismutase: An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 22: 6049-6055.
- Padmapriya V and N. Anand, 2009. Differential responses of superoxide dismutase in Anabaena variabilis Kütz to metal chelators. J. of Pure and Appl. Microbiol. Vol 3 (2).
- Forman, H. J. and I. Fridovich. 1973. Superoxide dismutase: a comparision of rate constants. Biochem. Biophys. 158: 396-400.







