Manuscript accepted on :24-Aug-2018
Published online on: 22-11-2018
Plagiarism Check: Yes
Reviewed by: Ajay Somkuwar
Second Review by: Neelapala Anil Kumar
Final Approval by: Prof. Alessandro Leite Cavalcanti
C. Anjanappa1 and H. S. Sheshadri2
and H. S. Sheshadri2
1Department of Electronics and Communication Engineering, The National Institute of Engineering, Mysore, Karnataka, India.
2Department of Electronics and Communication Engineering, PES College of Engineering, Mandya, Karnataka, India.
Corresponding Author E-mail: anjanappagayathri@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1589
Abstract
For proper modelling of signal and noise in MR data requires proper interpretation and analysis of data, the different approaches with this degradation due to random fluctuations in the MR data, probabilistic modeling is power solution, which needs correctness in the computation of noise is challenging task and various stastical approaches can be utilized. After modelling the noise it can be integrated to denoising pipeline, in this research work, the recognition of noise only pixels and the evaluation of standard deviation of noise using median, mean or other optimal sample quantiles are combined in to single frame work for noise assement and uses fixed point iterative procedure to obtain standard deviation of noise. We tested the effectiveness of the algorithm to the MR clinical and synthetic data base.
Keywords
Gamma Distribution; Noise Estimation; Parallel Reconstruction Algorithm; Rayleigh Distribution
Download this article as:| Copy the following to cite this article: Anjanappa C, Sheshadri H. S. Probabilistic Identification and Estimation of Noise: Application to MR Images. Biomed Pharmacol J 2018;11(4). |
| Copy the following to cite this URL: Anjanappa C, Sheshadri H. S. Probabilistic Identification and Estimation of Noise: Application to MR Images. Biomed Pharmacol J 2018;11(4). Available from: http://biomedpharmajournal.org/?p=24105 |
Introduction
Noise assement in MRI 1,2,3 usually requires the assessment of standard deviation of noise. Earlier methods can be separated into two methods for the computation of noise, first method involves the manually selected region of interest (ROI), in the second method entire volumetric data or image is considered for estimation without human interpretation. The problem is facing for the current automatic estimation method is separation of original signal from noise effected signals and other in homogeneity artifacts4,5 the proposed work of two authors is by aggregate the values of all pixels from an entire pixel data set in to one dimensional array and estimate the standard deviation of noise from the histogram of one dimensional array.in this research work, introduce simpler method for noise assement in MR image to eliminate the drawback of two authors. The proposed method requires the recognition of noise only pixels which enhance the performance and accuracy of estimation of Gaussian noise and requires the prior knowledge about the standard deviation of Gaussian noise, and can cater distribution of noise for further analysis like segmentation and registration. Most of the noise estimation algorithms in the earlier method is based on background methods and object based methods, background methods which follow Rayleigh distributed noise and object based methods follow Gaussian and rician noise.6 In this work we developed an algorithm for noise assement in MRI for the data acquired for multiple N receiver coils in which data follows nc- distribution is the special case of Rician distribution.7 Recently the author Rajan7,11 developed an algorithm for the estimation of noise without background. Most of the above estimation algorithms are developed are sensitive to artifacts results in inaccuracy of noise estimation, the author coupe8 developed robust noise estimation algorithm rician median absolute deviation [RMAD] for noise bias correction and artifacts to achieve accurate noise estimation, Other methods of noise estimation is variance stabilization transformation framework (VST) for bias inhomogeneity12,13 correction in MR image to transform bias Rician noise distribution in to uniform Gaussian distribution and achieves accuracy noise estimation compared to other noise estimation methods.
Materials and Method
The data set obtained from the DIPY (diffusion imaging in python) and JSS hospital Mysore Karnataka, India.
Data set 1
STANFORD_HARDI (High resolution Diffusion Weighted imaging dataset (N=4)) Uses phased array coil system and SOS reconstruction (without parallel imaging).
TAIWAN_NTU_DSI (Diffusion Spectrum imaging) Dataset (N=1) uses phased array coil system and SENSE reconstruction. (With parallel imaging).
Dataset 2
Philips 3.0 Tscanner
MRI T1 weighted axial Brain image having pathology acquisition parameters are TR=5.3sec, TE=20ms, slice thickness=3.5mm, Resolution of 512×512. Parallel Image Reconstruction: SENSE.
Philips 3.0Tscanner
MRI T1 weighted sagittal Brain image with acquisition parameters are TR=5.3 sec, TE=20ms, slice thickness=3.5mm, Resolution of 512×512. Parallel Image Reconstruction: SENSE.
Diffusion Imaging in Python (Dipy) is a free and open source software project tool where image processing library tools are available for the analysis of data from diffusion magnetic resonance imaging (dMRI) experiments.
Method
Noise modelling in MRI
The MR data acquired from the multiple N receiver coils m (or mi j k) follows non-central chi distribution [10] which is reconstructed from sum-of-squares algorithm (SOS) distribution,
![]()
distribution of 2N degrees of freedom with the non- centrality distribution parameter is given by
η2 /σg2. The probability density function (PDF) of
![]()
is given by [9]
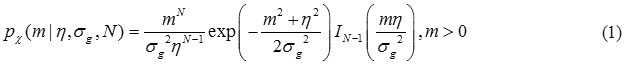
Where the PDF is zero for m < 0, η is the underlying (combined) signal intensity, σg is the standard deviation of Gaussian noise and Ik is the kth-order modified Bessel function of the first kind. We should note that magnitude MR signals reconstructed from other parallel image reconstruction techniques may not follow non-Central Chi distribution, Note that when N= 1, Eq. (1) reduces to the Rician PDF [2].
In this research work, the PDF of magnitude noise (i.e. η =0) is more relevant than the PDF of magnitude signal (η ≠ 0). Therefore, we will derive the PDF of magnitude noise from Eq. (1) by setting the input signal to zero, i.e. η =0, then it shows that PDF of magnitude noise is given by
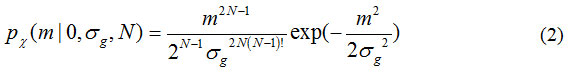
After IN-1 in Eq. (1) is replaced by first term Taylor expansion about input signal η =0,
Expressed by the following expression
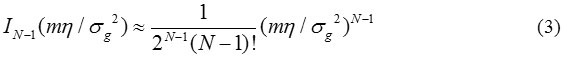
When N=1, Eq. (2) simplifies to Rayleigh PDF.
Then by a change of variables from m to
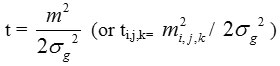
shows that variable t follows form of the particular Gamma PDF
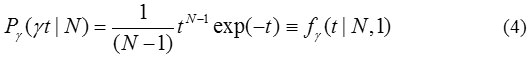
The Gamma PDF, fy as defined in [31]:
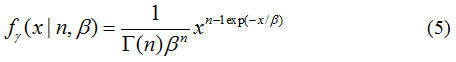
and Γ is the Gamma function.
Distribution of the Mean of Several Sampling Observed M-Values
The measurements m i, j, k’s are defined in Eq. (2). We have chosen the arithmetic mean for the identification of noise-only pixels. If knows the distribution for the mean, which is depends on σg, we can decide for any observed mean sample came from the noise-only distribution. Which will provide the expression for the arithmetic mean of the several sampling observed values of m i, j, k s through the method of Characteristic function. In short, the derived random variable si, j representing the arithmetic mean of K independent Gamma Random variables {ti,j,1, ti,j,2,…. ti,j,k} is given by
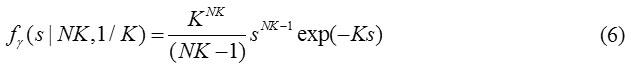
Further, the new random variable si ,j related to K independent magnitude MR measurements, {mi,j,1, mi,j,2, mi,j,k} drawn from the distribution of magnitude noise is given by
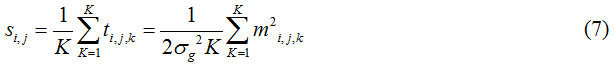
Identification of the noise-only pixels using probabilistic method requires to define threshold on ‘s’ i,e upper and lower threshold, so that a given proportion of all noise-only voxels or pixels fall between these two values. And we need to derive the cumulative distribution function (CDF), and it’s inverse of the distribution of ‘s’ to specify the lower and upper threshold values of s. Assume that N and K are known and the initial estimate of σg, is required, is obtained by automatically using the simple search method are below.
The probability distribution function and CDF of ‘s’ is denoted by
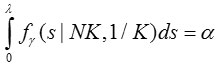
the inverse of CDF ‘s’ are denoted symbolically by λ= ps -1 (a | N, K).
The CDF of ‘s’ and its inverse are given by
λ= ps -1 (ps ( λ| N, K).| N, K) and a = ps-1 (ps ( a| N, K)| N, K) Note that Ps-1 (a|N,K) given by Inverse Gamma Regularized [NK,1-α]|K in Mathematica[14].The lower and upper threshold values of ‘s’ are λ –, λ + denoted probabilities are a/2 and 1-(a/2) can be expressed in terms of the inverse CDF of ‘s’ given by λ – = Ps-1 (a/2|N,K), and λ + Ps-1 (1-a/2|N,K), Thus, a collection of K independent magnitude MR signals {m i,j,1, m i,j,2, m i,j,k} from the acquisition is decided to contain only noise if
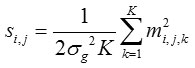
which satisfies the inequalities, λ – ≤ S i,j, ≤ λ +.
Noise Estimation using the sample median, the sample mean or optimal quantiles method
The computation of Gaussian noise SD is obtained from the simple method of mean, denoted by <m>, of means of a collection of acquisition data measurements. The estimation of Gaussian noise SD, σg, can also be computed from the sample quantile of a unique optimal order α*
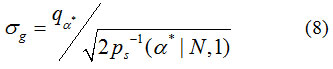
Note that both the sample median and the median of the continuous PDF, Eq. (2), are denoted by the same symbol. Note also that the denominator of Eq. (9) can be computed in advance, see Table 2. For example, when N = 1, Eq. (9) can be expressed mathematically
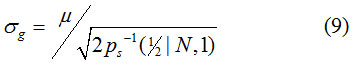
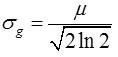
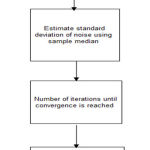 |
Figure 1: Block diagram of PIESNO Algorithm.
|
Algorithm Implementation
Input parameters: N, K, λ –, λ+, m i,j,k for all input measurements, the estimate of σg, is needed
Output framework: identification of noisy only pixels and estimate of σg, and array of noise elements ‘Ω’
Step 1 is the identification of noise-only pixels, and dynamically adding noisy pixels in to new elements of the one-dimensional array, Ω. And compute the standard deviation of noise in advance using the equation shown below.
At each pixel location (i,j), compute si,j:
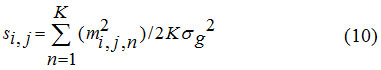
A positive identification is made if S i,j satisfies the condition λ – ≤ S i,j, ≤ λ +.
In Step 2, the channel between the identification and estimation of noise-only pixels is set and create one-dimensional array of Ω, which is the union of all the arrays of K measurements identified as noise only pixels. Note that the number of positive identifications is defined is equal to the number of the positively identified arrays. Finally, the sample median of Ω, 1, is selected. Note again that the sample mean of Ω or the sample quantile of Ω of a specific order may be used here.
Step 3 is the computation of standard deviation of noise, the array of sample median, l, of Ω computed in Step 2 is used to estimate σg. Note again that the sample mean and other optimal quantiles are defined below.
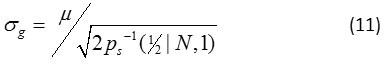
In Step 4, if the iteration stops the Ω is empty, which repeats procedure from Steps 1 to 4 sequentially until convergence or the maximum number of iterations is reached. Here, a sequence of estimates of σg produced by the iterative procedure is considered if the absolute difference between any successive estimates is less than small number, say 0.00000000001.
Step 5, the noise identification and computation of the standard deviation of noise is denoised using NLM algorithm and evaluate its performance parameters.
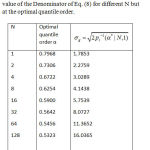 |
Table 1: The optimal quantile value for different N and the value of the Denominator of Eq. (8) for different N but at the optimal quantile order.
|
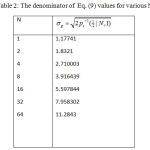 |
Table 2: The denominator of Eq. (9) values for various N.
|
Results and Discussion
The proposed algorithm is implemented using Python. After the python implementation of the PIE S. NO we plot the SD of noise-only pixels using MATPLOTLIB with N (phased array coils) varying from 1 to 12. The figure (4-6) below shows the plots of PDF of noise-only pixels vs SD (sigma) with N=1, 4 and 10. Using different data sets below.
Piesno Results for Different Datasets
Tested on Taiwan and Stand ford Dataset
Fig.2 and fig.3 shows the data set obtained from Stanford_hardi dataset acquired using 4 phased array coils. In these images the White dots appearing in the image are the noise-only pixels identified and estimated using our proposed PIESNO algorithm. These noisy only pixels are denoised using well known NLM algorithm using python. The image is subjected to denoising with Background preservation. Similarly the data set obtained from Taiwan dataset acquired using single phase array coil, from these two graphs we observed that the noise identified is Rician and Gaussian from the background region and it is well modelled by Gaussian and rician from the PDF curve is as shown in fig.4 and fig.5.
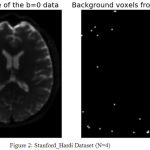 |
Figure 2: Stanford_Hardi Dataset (N=4).
|
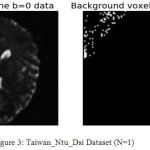 |
Figure 3: Taiwan_Ntu_Dsi Dataset (N=1).
|
 |
Figure 4: N=1 Rician distribution.
|
From the algorithm the estimated value of standard deviation of noise and its PDF curve is shown in fig.4 above. The curve is not symmetric about a mean value. Hence we identify the noise present in the MRI as Rician Noise or nc-λ distribution.
 |
Figure 5: N=4 Gaussian distribution.
|
For the fig.4 above estimated value of standard deviation of noise using PIESNO algorithm using N=4, the plot obtained is a symmetric curve as shown above. The curve is symmetric about the value 0. Hence it follows a normal distribution and the noise is identified as a Gaussian Noise.
Similarly for N>1, the plot follows the same curve as shown below fig.5 for N=10.
 |
Figure 6: N=10 Gaussian distribution.
|
Thus we can interpret the results of PIESNO as
For N=1 the noise estimated and identified is Rician Noise.
For N>1 the noise estimated and identified is Gaussian Noise.
Tested on Clinical Data
Fig.7 shows the data acquired from MR machine (clinical data) using multiple phase array coils using parallel image reconstruction SENSE algorithm and fig. 8 shows data acquired using SOS reconstruction algorithm without subsampling. The identification of noise distribution using PIESONO algorithm and its results with histograms is as shown in fig. 9-12, from this graphical analysis found that the noise distribution is Gaussian and rician.
The denoising results of the algorithm for Taiwan and stand ford dataset are shown in fig.13 and fig.14 and difference image (Residual image) is shown, it is observed that the algorithm shows better preservation of edge and efficient noise reduction.
The comparison of denoising algorithm with other algorithm is shown in table 3. From this table it is observed that the NLM algorithm shows efficient noise reduction with others and it is verified from PSNR values, and it is burden due to long execution time compared with AONLM and PCA algorithm is shown in the table 3.
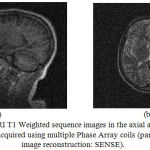 |
Figure 7: MRI T1 Weighted sequence images in the axial and sagittal view acquired using multiple Phase Array coils (parallel image reconstruction: SENSE).
|
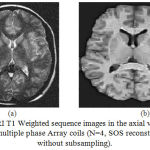 |
Figure 8: MRI T1 Weighted sequence images in the axial view acquired using multiple phase Array coils (N=4, SOS reconstruction without subsampling).
|
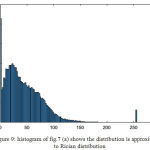 |
Figure 9: Histogram of fig.7 (a) shows the distribution is approximated to Rician distribution.
|
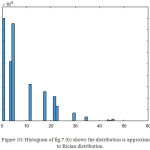 |
Figure 10: Histogram of fig.7 (b) shows the distribution is approximated to Rician distribution.
|
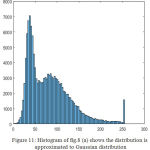 |
Figure 11: Histogram of fig.8 (a) shows the distribution is approximated to Gaussian distribution.
|
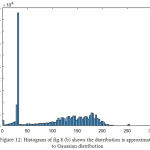 |
Figure 12: Histogram of fig. 8 (b) shows the distribution is approximated to Gaussian distribution.
|
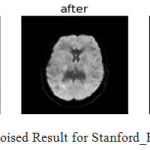 |
Figure 13: Denoised Result for Stanford_Hardi (N=4).
|
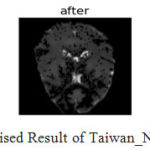 |
Figure 14: Denoised Result of Taiwan_Ntu_Dsi (N=4).
|
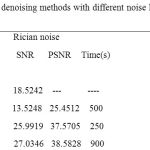 |
Table 3: The comparison of denoising methods with different noise levels on MR T1 weighted Images.
|
Discussion
Earlier estimation algorithms work for single image but our proposed algorithm is well suited for multiple images to estimate the variance of noise and it is not suited for clinical practice requires longer acquisition time to estimate the level of noise. Our method is applicable to larger class of magnitude data measurements acquired from the MR machine than Rayleigh-distributed data (i.e., N = 1) and discuss some limitations of the proposed approach. PIESNO cannot be expected to perform well for very small K because the identification of noise may be less stable and may not produce sufficient number of elements for the estimation of noise via the median method to be useful as an estimator. We tested the effectiveness of the algorithm with clinical and synthetic database and conclude that stastical distribution of noise is Gaussian, rician or nc-λ distribution and it is verified from mathematical derived probabilistic distribution function. The denoising performance of the algorithm is compared with standard conventional algorithm and execution time.
References
- Macovski. Noise in MRI, Magn. Reson. Med. 1996;36:494–497.
CrossRef - Gudbjartsson H.,Patz S. The Rician distribution of noisy MRI data.Magnetic Resonance in Medicine. 1995;34(6)910–914.
CrossRef - Drumheller. General expressions for Rician density and distribution functions. IEEE Trans. on Aerospace and Electronic Systems. 1993;29(2):580–588.
CrossRef - Sijbers., den Dekker J. A., Van Verhoye A. M.,Van Dyck D. Estimation of the noise in magnitude MR images. Magn. Reson.Imag. 1998;16(1):87–90.
- Sijbers., Poot D., den Dekker A. J and Pintjens W. Automatic estimation of the noise variance from the histogram of a magnetic resonance image. Physics in Medicine and Biology. 2007;52(5):1335-1348.
CrossRef - Aja-Fernandez., Niethammer M., Kubicki M., Shenton M. E and Westin C. F. Restoration of dwi data using a rician lmmse estimator. Medical Imaging. IEEE Transactions on. 2008;27(10):1389-1403.
- Aja-Fernandez., Alberola-Lopez C and Westin C. F. Noise and signal estimation in magnitude MRI and Rician distributed images: a LMMSE approach. IEEE transactions on image processing. 2008;17(8):1383-1398.
CrossRef - Sijbers., den Dekker A., Scheunders P.,Van Dyck D. Maximum-likelihood estimation of Rician distribution parameters. IEEE Trans. Med.Imaging. 1998;17(3):357–361.
CrossRef - Koay G., Basser P. J. Analytically exact correction scheme for signal extraction from noisy magnitude MR signals.Journal of Magnetic Resonance. 2006;179(2):317–322.
CrossRef - Aja-Fernandez., Tristan-Vega A., Alberola-Lopez C. Noise estimation in single- and multiple-coil magnetic resonance data based on ´statistical models. Magn. Reson. Imag. 200;27:1397–1409.
- Rajan., Poot D., Juntu J., Sijbers J. Noise measurement from magnitude MRI using local estimates of variance and skewness. Physics in Medicine and Biology. 2010;55(16):441.
CrossRef - Coupé P., Manjón J., Gedamu E., Arnold D., Robles M and Collins D. L. Robust Rician noise estimation for MR images. Medical Image Analysis. 2010;14(4):483–493.
CrossRef - Foi A. Noise estimation and removal in MR imaging: the variance-stabilization approach,” in [Proceedings of the IEEE International Symposium on Biomedical Imaging: From Nano to Macro]. 2011.
CrossRef - Wolfram S. The Mathematica Book, Wolfram Media, Champaign. 2003.







