Manuscript accepted on :26 June 2018
Published online on: 28-06-2018
Plagiarism Check: Yes
Anjali Devi S. Bettadapura1, Vijay K. Munivenkatappa2 and Subba Rao V. Madhunapantula1,3
1Department of Biochemistry, JSS Medical College, JSS Academy of Higher Education and Research, Mysuru - 570015, Karnataka, India.
2Department of Oncosurgery, Bharath Hospital and Institute of Oncology, Mysuru - 570 017, Karnataka, India.
3Special Interest Group in Cancer Biology and Cancer Stem Cells (SIG-CBCSC), JSS Academy of Higher Education and Research, Mysuru - 570015, Karnataka, India.
Corresponding Author E-mail: madhunapantulas@yahoo.com
DOI : https://dx.doi.org/10.13005/bpj/1472
Abstract
Carcinoma of cervix is one of the major health concerns among women. Although Effective preventive methods are available to protect individuals from getting cervical cancer, the incidence and mortality rates due to this disease are increasing at alarming rates. In addition, currently, no potent treatment strategies exist to treat advanced cervical cancers. Therefore, identification of key biomarkers that indicate the severity and drug resistance of cervical cancers is of high importance. Comparative analysis of transcriptome of tumor- and normal tissue is one way of determining the deregulated genes, which serve as biomarkers. Hence, in this study the transcriptome of malignant cervical cancer tissues was compared with adjacent non-tumor tissue and genes that are at least 2-fold high were selected. Among various selected genes, Bone gamma carboxyglutamic acid containing protein (BGLAP, commonly known as Osteocalcin) was found consistently high in all the tumor tissue compared to adjacent non-tumor tissue. Hence, BGLAP could be a potential biomarker for identifying malignant cervical cancer tumors.
Keywords
BGLAP; Carboxyglutamic; Effective
Download this article as:| Copy the following to cite this article: Bettadapura A. D. S, Munivenkatappa V. K, Madhunapantula S. R. V. Transcriptome Analysis Identified Elevated Expression of Bone Gamma-carboxyglutamic acid-containing Protein (BGLAP) in Human Cervical Cancer Tissues. Biomed Pharmacol J 2018;11(2). |
| Copy the following to cite this URL: Bettadapura A. D. S, Munivenkatappa V. K, Madhunapantula S. R. V. Transcriptome Analysis Identified Elevated Expression of Bone Gamma-carboxyglutamic acid-containing Protein (BGLAP) in Human Cervical Cancer Tissues. Biomed Pharmacol J 2018;11(2). Available from: http://biomedpharmajournal.org/?p=21172 |
Introduction
Cervical cancer is one of the major health concern among women globally. Even though the incidence of cervical cancer is on a declining trend in developed countries due to effective screening and vaccination, the mortality rate is still about 30%.1 According to 2016 United States statistics about 12990 new cases of carcinomas of uterine cervix are estimated to be diagnosed of which 4120 cases result in death.1 In sub Saharan Africa about 75000 new cases of cervical cancer have been detected every year resulting in 55000 deaths. World Health Organisation has estimated that about 4,43,000 deaths will occur from cervical cancer by 2030 world wide.2 In Latin America cervical cancer continues to be the second leading cause of deaths in women.3 For example, in the year 2015, an about 74488 women were diagnosed with cervical cancer of which 31003 cases were succumbed to death.3 Despite several screening and vaccination programs, it is predicted that the mortality from cervical cancer will increase to ~45% by 2030.3 According to national cancer registry, in India, approximately 113,138 new cervical cancer cases were detected in the year 2015 making it the third most common cancer.4 Further analysis showed that about a quarter of cervical cancers diagnosed each year are from India.4 Recent projections predicted an increase of up to 123,291 cases by the year 2020.4 Therefore, the burden of cervical cancer continues to increase, especially in developing and under developed countries as it affects women in the productive age group of 20 to 59 years.5,6 Early detection is one of the ways to reduce the mortality rates, and transcriptome analysis is an approach widely used to identify deregulated genes in cancer cells. Comparison of the transcriptomes of non-tumor and tumor tissues provide key information about the genes that are deregulated in tumors, hence, is a useful method to identify biomarkers of tumor progression and disease severity.
Transcriptome is the entire collection of genes that are transcribed in a particular cell, and is an indicator of gene expression.7 In general, transcriptome is analyzed by, first, isolating the total RNA followed by separating the mRNA and measuring the level of expression based on the number of RNA transcripts.7.8 Sequencing of mRNA transcripts provide information even about the mutations present in genes.7,8 Therefore, transcriptome analysis is an important tool not only to measure gene expression but also to identify genetic changes in a cell.7,8 Hence, in this study the transcriptome analysis was carried out to determine the key genes that are deregulated in cervical cancer tissues compared to paired non-tumor tissue. One deregulated gene identified in this analysis is bone gamma-carboxyglutamic acid-containing protein (BGLAP).9
BGLAP, also known as a Osteocalcin, is a 11kDa protein hormone synthesized and secreted by osteoblasts.10 Functionally, it regulates the mineralization of bone matrix and control coagulation of blood.11,12 However, recent studies have implicated BGLAP in tumor progression and malignant transformation of cells.13.14 For instance, elevated BGLAP was reported in carcinomas of prostate and breast, and has been shown to promote metastasis of these tumor cells to bone.13.14 Similarly, a recent study evaluating the role of BGLAP in pancreatic cancer demonstrated a vital function to this protein in the survival and invasion of cells.15 However, to date no study has reported the expression of BGLAP in cervical cancers. To the best of our knowledge, this is the first report mentioning the elevated expression of BGLAP in carcinomas of cervix. Future studies should evaluate the mechanisms by which BGLAP is promoting the growth of cervical cancer cells and whether targeted inhibition of BGLAP helps in retarding survival and spread of cells.
Materials and Methods
Collection of Tissues
Ethical committee clearance was taken before the commencement of the study (JSS/MC/IEC/4898/2012-2013 dated 22-12-2012). Paired cancerous- and adjacent non-cancerous tissues were collected from patients visiting the Department of Oncosurgery, Bharath Hospital and Institute of Oncology (a collaborative hospital located in Mysore) and JSS Hospital after obtaining informed consent. The samples were collected by punch biopsy. The study participants were not exposed to any form of cancer therapy. The collected tissues were immediately transferred to DNAse and RNAse free tubes containing RNA later solution (~500µL). The collected tissues were stored at -200C freezer.
Extraction of total RNA
RNA extraction was carried out using Allprep RNA extraction kit from Qiagen India Pvt Ltd, New Delhi, India (Scheme #1). The kit extracts all RNA molecules that are longer than 200 nucleotides.16 Procedurally, first, the tissues (30.0mg) were lysed and homogenized in a high-density guanidino isothiocyanate containing buffer to inactivate DNAses, RNAses and proteases. Next, the lysate was passed through DNA spin column, which allows the binding of genomic DNA. The column was washed thrice with wash buffer, and the bound DNA eluted. After this, the sample free of DNA was made to flow through the RNeasy spin column, which facilitates the binding of total RNA to the column. The contaminants and impurities were washed and the bound RNA eluted using 30µL water.
Determination of the quality of isolated RNA
Quality of isolated RNA was assessed using tape-station. Principally, the tape-station works similar to agarose gel electrophoresis.17 Samples were prepared by adding 5.0µL buffer to 1.0µL RNA. Since RNAs possess tertiary structures, the samples were denatured by heating at 72oC for 2 minutes. The denatured samples were placed on ice (for 2 minutes) and quality scores determined by assessing RNA Integration Number (RIN). RIN is a software algorithm developed by Agilent technologies to analyze the quality of RNA. It considers the whole electrophoretogram into account before ascertaining an integer value. RIN ranges from 1 to 10, with 10 being the most intact and 1 being the most degraded RNA.17 Samples with RIN equal to or greater than 7 were used for library preparation in this study.
Quantification of Isolated RNA Using Qubit Method
The amount of RNA present in the sample was determined using RNA HS assay kit fluorimetrically. Qubit RNA hS kit accurately determines the RNA concentration over a range of 250 pg/ml to 100ng/ml. Experimentally, first, the Qubit RNA HS reagent was diluted 1: 200 in Qubit RNA buffer solution. Next, the concentration curve was prepared by mixing 10.0µL standards with 190.0µL working solution. The samples (1.0 to 20.0µL) were prepared by adjusting the final volume to 200µL using Qubit working solution. All the tubes were incubated at room temperature for two minutes and the values read using Qubit fluorimeter operating with excitation filters: Blue 430–495nm; and Red 600–645nm; emission filters: Green 510–580nm, and Red 665–720nm.
Preparation of libraries
Step#1: Purification of mRNA and generation of fragments: mRNA from the total RNA was purified using magnetic beads that contain Oligo dT. Bound RNA was eluted by the addition of polyA RNA.18 Fragmentation of RNA was carried out during the elution of RNA from beads. Next, the primers for cDNA synthesis were added to the eluted fragmented RNA and the synthesized cDNA subjected for (a) end repair protocol to remove 31 overhangs, and (b) filling the gaps in 51 overhangs using polymerase.19 Next, the 3’ ends were adenylated by adding a single Adenylate nucleotide, which prevents the relegation.19 Next, the adapters were ligated and analyzed by flow cell hybridization. The adapters utilized are
TruSeq Universal Adapter: 5’AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT3’
TruSeq Illumina Index Adapter:
5’GATCGGAAGAGCACACGTCTGAACTCCAGTCAC[INDEX]ATCTCGTA TGCCGTCTTCTGCTTG3’
DNA Fragment Enrichment
PCR was used to amplify DNA fragments
The program conditions used were detailed below:
Initial denaturation – 98°C for 30 seconds
PCR cycles (15 cycles):
Denaturation – 98°C for 10seconds; Annealing – 60°C for 30seconds,
Elongation: 72°C for 30seconds
Final extension: 72°C for 5 minutes
Storage: Hold at 10°C
After PCR amplification, the prepared library was quantified using Qubit DNA assay reagent kit (concentration range of 10.0pg/µL to 100.0ng/µL), and the size of DNA fragments determined using Tape station-2200, which has been designed to analyze DNA of size 35-1000bp
Normalization of library
DNA templates were prepared ready for cluster generation as detailed in.20,21
Cluster generation
The cluster generation was carried out by amplifying the library on a cBOT using solid phase PCR, (on a flow cell coated), with oligonucleotide probes. The dsDNA was denatured to single strands to enable the binding of primers for sequencing. The amplified libraries were sequenced using sequencing by synthesis method on HiSeq 4000 using 100bp paired end chemistry.
Analysis of Sequenced Data
The data acquired from fast-Q files (obtained from HiSeq 4000), was trimmed using the trimmomatic software version 0.036. Procedurally, the trimming protocol includes: (a) Removal of adaptors and other illumina specific sequences using Illumina Clip; (b) Trimming of reads with below the average quality by scanning from the 5’ end; (c) Maximization of the quality of each read by balancing error rate and read length; (d) Removal of bases at the start, and end of the read respectively if the quality is below the threshold; (e) Elimination of bases from the start and end of the read; (f) Dropping of the reads of below minimum length; (g) Removal of reads below average quality; (h) Convert quality scores to Phred 33 and Phred 64.
Contamination Removal
Removal of Non mRNA and mitochondrial genome sequences The unwanted sequences that include – mitochondrial genome sequences, ribosomal RNAs, transfer RNAs, adapter sequences and others were removed using Bowtie 2 Version 2.2.4
Read Alignment
The paired-end reads were aligned to the reference human genome Feb. 2009 release, which was downloaded from UCSC database (GRCh37/hg19). The chromosome fasta file was downloaded from the website and alignment was performed using Hierarchical Indexing for Spliced Alignment of Transcripts program.
Read Summarization
Read summarization of the samples was estimated using Feature Counts (version 1.5.2). These counts mapped reads for genes, exons, promoter, gene bodies, genomic bins and chromosomal locations and can be used for both RNA and DNA sequences.
Differential Expression
The Differential expression analysis was performed using DESeq 2. This program uses negative binomial distribution for analyzing differential gene expression (Scheme #1).
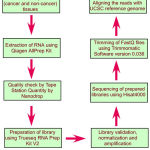 |
Scheme 1: Flow chart depicting different steps in the transcriptome analysis.
|
Results
Rna Isolated From Tissues Is Not Degraded And Is Suitable For Cdna Library Preparation
Total RNA from cervical cancer tissues as well as adjacent non-tumorous samples was isolated as detailed in materials and methods section and the quality and quantity determined using nanodrop spectrophotometer and Tape Station (Figure 1 and Table 1). Analysis of the data showed the presence of high quality RNA as evidenced by 260/280 ratio more than 2.0 for majority of samples, except non-cancerous tissue #5 (NC5, 260/280 = 1.70) and cancer tissue (CT5, 260/280 = 1.66). In addition, the RNA integration number (RIN), an indicator of RNA quality, is more than 7.0 for all the samples except cancer tissue #5 (CT5, RIN = 6.5) and non-cancerous tissue #1 (NC1, RIN = 6.8). Although RIN above 8.0 is considered of high quality, for tumor tissues values in the range of 6.0 to 8.0 are also considered suitable for library preparation (Table 1).
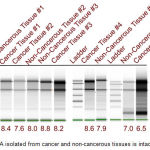 |
Figure 1: RNA isolated from cancer and non-cancerous tissues is intact.
|
In order to test the quality of RNA isolated from cervical cancer and adjacent non-cancer tissues the samples were analyzed by electrophoresis on Tape Station as detailed in materials and methods. Presence of sharp bands representing 18S and 28S rRNA indicates that the isolated RNA has not undergone any degradation, hence, is suitable for further processing. The RNA integration number (RIN number) (>6.5 for tissues) also indicates the suitability of isolated RNA for cDNA library preparation.
Table 1: Qualitative and quantitative analysis of isolated RNA
| Sample | A260/280 Ratio | A260/230 Ratio | QUBIT (ng/µl) | RIN |
| Non-cancerous tissue #1 | 2.02 | 1.17 | 40.2 | 6.8 |
| Non-cancerous tissue #2 | 2.07 | 2.14 | 414.0 | 8.0 |
| Non-cancerous tissue #3 | 2.07 | 2.09 | 496.0 | 8.8 |
| Non-cancerous tissue #4 | 2.13 | 0.53 | 72.0 | 7.9 |
| Non-cancerous tissue #5 | 1.70 | 0.46 | 110 (1:4) | 7.0 |
| Cancer tissue #1 | 2.07 | 0.84 | 80.0 | 8.4 |
| Cancer tissue #2 | 2.08 | 2.03 | 230.0 | 7.6 |
| Cancer tissue #3 | 2.08 | 1.47 | 998.0 | 8.2 |
| Cancer tissue #4 | 2.09 | 1.53 | 10.9 | 8.6 |
| Cancer tissue #5 | 1.66 | 0.32 | 398 | 6.5 |
BGLAP is upregulated in cervical cancer tissues compared to adjacent non-cancerous tissue
Since the RNA isolated from cervical cancer tissues as well as adjacent non-cancerous tissues is suitable for preparing the cDNA library, all the samples were processed for further processing as detailed in the Scheme #1. Analysis of the sequencing data followed by bioinformatics calculations identified BGLAP as one of the key genes upregulated constantly in all the samples (Figure 2A and 2B).
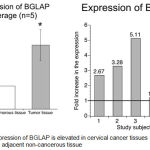 |
Figure 2: Expression of BGLAP is elevated in cervical cancer tissues compared to adjacent non-cancerous tissue.
|
Analysis of transcriptome data and the comparison of identified genes between adjacent non-cancer tissue and cancer tissue showed elevated expression of BGLAP in all the 5 paired samples indicating BGLAP could be a potent oncogene in cervical cancers. Average expression showed an about 2.7 fold increase in the mRNA expression compared to normal tissues.
Discussion
BGLAP the widely known Osteocalcin is a key oncogenic protein expressed by many metastatic tumors.22 Very recently Hayashi, Y et al., have demonstrated that osteocalcin regulates prostate cancer cells growth in a carboxylation dependent manner.23 For example, whereas the gammacarboxylated osteocalcin (GlaOC) promoted the proliferation of prostate cancer cells, the uncarboxylated osteocalcin retarded the cell growth by controlling the phosphorylation of receptor tyrosine kinases. 23 Therefore, it is important to, first, measure the expression of Osteocalcin in tumor cells compared to adjacent non-cancerous tissues. In this study we have shown elevated expression of BGLAP in all the cervical cancer tissues compared to adjacent non-cancerous tissues indicating this could be a potent oncogene in cervical cancers. However, it is not known whether BGLAP expression at mRNA level also correlate with protein expression in tissues as well as in serum, if so, the expressed BGLAP is functionally active. More over it is also currently unknown whether any polymorphic forms of BGLAP are present in cervical cancer tissues. A study by Liu, Y et al., showed a significant association between the rs1800247 HH and Hh genotypes and increased susceptibility to hepatocellular carcinoma.24
A separate study has demonstrated that BGLAP regulate tumor cell proliferation, survival and metastasis of pancreatic cells.15 Targeting knockdown BGLAP using siRNA or expressing TNF-a, a negative modulator of BGLAP in pancreatic cancers, reduced the proliferation and migration of pancreatic cancer cells.15 Hence, BGLAP is considered as a marker for tumor cell proliferation and migration.15
Although many prior studies have reported newer pathways in cervical cancers based on transcriptome analysis, to date, no study has identified BGLAP as a key marker for metastatic cervical cancers. For example, a study by Campos-Parra, A.D. et al., 2016 analyzing the transcriptomes of 89 locally advanced cervical cancer tissues has shown over expression of 7530 genes related to 93 signaling pathways such as JAK-STAT, NOTCH and mTOR autophagy cascades.25 Many other studies have also carried out transcriptomic analysis of cervical cancer tissues and identified unique pathways. Even though we have identified BGLAP in this study, it’s expression in multiple sample needs to be tested and validation of its involvement needs to be experimentally confirmed using proof-of-principle studies. Therefore, future studies should measure the expression of BGLAP in the serum and tissues of cervical cancer patients and test whether knocking down has any impact in cervical cancer cell growth.
Conclusions
In summary, we have carried out the analysis of transcritome of cervical cancer tissues and compared with adjacent non-cancerous tissues to identify key deregulated genes. Analysis of the data showed a consistent upregulation of BGLAP gene exclusively in cancer tissues but not in non-cancerous tissues, indicating it could be a potent therapeutic target in this cancer. However, additional studies are warranted to check its role in cervical cancer cells development, growth and metastasis.
References
- Siegel R.L, Miller K.D, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016Jan1;66(1):7-30.
- Mboumba R.B, Prazuck T, Lethu T, Meye J.F, Bélec L. Cervical cancer in sub-Saharan Africa: an emerging and preventable disease associated with oncogenic human papillomavirus. Medecine et sante tropicales. 2017 Feb;27(1):16-22.
- Bychkovsky B.L, Ferreyra M.E, Strasser‐Weippl K, Herold C.I, de Lima Lopes G, Dizon D.S, Schmeler K.M, Del Carmen M, Randall T.C, Nogueira‐Rodrigues A, de Carvalho Calabrich A.F. Cervical cancer control in Latin America: a call to action. Cancer. 2016 Feb15;122(4):502-14.
CrossRef - Takiar R, Nadayil D, Nandakumar A. Projections of number of cancer cases in India (2010-2020) by cancer groups. Asian Pac .J. Cancer Prev. 2010 Jan1;11(4):1045-9.
- Sreedevi A, Javed R, Dinesh A. Epidemiology of cervical cancer with special focus on India. International journal of women’s health. 2015;7:405.
- Mallath M.K, Taylor D.G, Badwe R.A, Rath G.K, Shanta V, Pramesh C.S, Digumarti R, Sebastian P, Borthakur B.B, Kalwar A, Kapoor S. The growing burden of cancer in India: epidemiology and social context. The Lancet Oncology. 2014 May 1;15(6):e205-12.
CrossRef - Wolf J.B. Principles of transcriptome analysis and gene expression quantification: an RNA‐seq tutorial. Molecular ecology resources. 2013 Jul 1;13(4):559-72.
CrossRef - Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T. Transcriptomics technologies. PLoS computational biology. 2017 May18;13(5):e1005457.
CrossRef - Price P.A, Otsuka A.A, Poser J.W, Kristaponis J, Raman N. Characterization of a gamma-carboxyglutamic acid-containing protein from bone. Proceedings of the National Academy of Sciences. 1976 May 1;73(5):1447-51.
CrossRef - Puchacz E, Lian J.B, Stein G.S, Wozney J, Huebner K, Croce C. Chromosomal localization of the human osteocalcin gene. Endocrinology. 1989 May1;124(5):2648-50.
CrossRef - Zoch M.L, Clemens T.L, Riddle R.C. New insights into the biology of osteocalcin. Bone. 2016 Jan1;82:42-9.
CrossRef - Hauschka P.V. Osteocalcin: the vitamin K-dependent Ca2+-binding protein of bone matrix. Pathophysiology of Haemostasis and Thrombosis. 1986;16(3-4):258-72.
CrossRef - Stracke H, Schatz C, Pralle H, Ullmann J, Schatz H. Osteocalcin, a marker in diseases with elevated bone metabolism. Deutsche medizinische Wochenschrift (1946). 1985 Sep;110(38):1442-6.
- Thulin M.H, Jennbacken K, Damber J.E, Welén K. Osteoblasts stimulate the osteogenic and metastatic progression of castration-resistant prostate cancer in a novel model for in vitro and in vivo studies. Clinical & experimental metastasis. 2014 Mar 1;31(3):269-83.
CrossRef - Kayed H, Bekasi S, Keleg S, Michalski C.W, Giese T, Friess H, Kleeff J. BGLAP is expressed in pancreatic cancer cells and increases their growth and invasion. Molecular cancer. 2007 Dec;6(1):83.
CrossRef - Qiagen G. RNeasy® Mini Handbook.
- Padmanaban A, Salowsky R, Cher C. RNA quality control using the agilent 2200 TapeStation system–assessment of the RIN e quality metric. Agilent technologies application notes. 2012.
- Peng G, Dan W, Jun W, Junjun Y, Tong R, Baoli Z, Yang X. Transcriptome profiling of the cancer and adjacent nontumor tissues from cervical squamous cell carcinoma patients by RNA sequencing. Tumor Biology. 2015 May 1;36(5):3309-17.
CrossRef - Trombetta J.J, Gennert D, Lu D, Satija R, Shalek A.K, Regev A. Preparation of Single‐Cell RNA‐Seq Libraries for Next Generation Sequencing. Current protocols in molecular biology. 2014 Jul 1:4-22.
- Kumar R, Ichihashi Y, Kimura S, Chitwood D.H, Headland L.R, Peng J, Maloof J.N, Sinha N.R. A high-throughput method for Illumina RNA-Seq library preparation. Frontiers in plant science. 2012 Aug 28;3:202.
CrossRef - Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols. 2010 Jun 1;2010(6):pdb-rot5448.
- Nordstrand A, Bovinder Ylitalo E, Thysell E, Jernberg E, Crnalic S, Widmark A, Bergh A, Lerner U.H, Wikström P. Bone Cell Activity in Clinical Prostate Cancer Bone Metastasis and Its Inverse Relation to Tumor Cell Androgen Receptor Activity. International journal of molecular sciences. 2018 Apr 18;19(4):1223.
CrossRef - Hayashi Y, Kawakubo-Yasukochi T, Mizokami A, Hazekawa M, Yakura T, Naito M, Takeuchi H, Nakamura S, Hirata M. Uncarboxylated Osteocalcin Induces Antitumor Immunity against Mouse Melanoma Cell Growth. Journal of Cancer. 2017;8(13):2478.
CrossRef - Liu Y, Huang L, Lu Y, Xi X.E, Huang X.L, Lu Q, Huang X, Li S, Qin X. Relationships between the osteocalcin gene polymorphisms, serum osteocalcin levels, and hepatitis B virus-related hepatocellular carcinoma in a chinese population. PloS one. 2015 Jan 14;10(1):e0116479.
CrossRef - Campos-Parra A.D, Padua-Bracho A, Pedroza-Torres A, Figueroa-González G, Fernández-Retana J, Millan-Catalan O, Peralta-Zaragoza O, de León D.C, Herrera L.A, Pérez-Plasencia C. Comprehensive transcriptome analysis identifies pathways with therapeutic potential in locally advanced cervical cancer. Gynecologic oncology. 2016 Nov 1;143(2):406-13.
CrossRef







