Manuscript accepted on :28 April 2018
Published online on: 11-05-2018
Plagiarism Check: Yes
Maysaa Kadhim Al-Malkey1, Zainab Abdul Jabbar Aldhaher2, Rasha Abbas Azeez2, Sumaiah Ibrahim Hussein2, Sinai Waleed Mohammed1, Munira Ch Ismeeal3 and Khawla Ibrahim Mesheal1
1Tropical-Biological Research Unit, College of Science, University of Baghdad, Baghdad-Iraq.
2Department of Basic Sciences, College of Dentistry, University of Baghdad, Baghdad-Iraq.
3Department of Microbiology, College of Science, Al-Karhk University of Science, Baghdad-Iraq.
Coressponding Author E-mail: maysakadhim@uobaghdad.edu.iq
DOI : https://dx.doi.org/10.13005/bpj/1426
Abstract
Oral squamous cell carcinoma (OSCC) is the most common malignant neoplasm of the oral mucosa. Human papillomavirus (HPV) virus cause a broad scope of diseases from benign to invasive tumors, types 16 and 18 classified as carcinogenic to humans. This study aimed to provide the first molecular characterization of HPV types in Iraq. Thirty-five unstimulated whole saliva samples were collected from histopathologically confirmed patients with oral cancer were enrolled in this study. Genomic DNA was extracted from exfoliating cells to amplify HPV-DNA using HPV-L1 gene sequence primers by polymerase chain reaction method (PCR), the viral genotyping was performed using direct sequencing method. HPV genotypes identified were deposited in GenBank. HPV DNA was detected in 20 of 35 OSCC patients representing (57%).The most frequent HPV genotypes were HPV-18 accounting for (75%) (15 out of 20 patients) followed by HPV-16 accounting for (20%) (4 out of 20), and HPV-11 accounting for (5%) (5 out of 20 patients). This study highlights the high-risk HPV genotypes in OSCC patients and their phylogenetic analysis tree and their homology to the ancestral sequence which may indicate emerging of a new biological entity of HPV-positive OSCC with a potential sexually transmission.
Keywords
HPV; L1 Gene; OSCC; Phylogenetic Tree
Download this article as:| Copy the following to cite this article: Al-Malkey M. K, Aldhaher Z. A, Azeez R. A, Hussein S. I, Ismeeal M. C, Mohammed S. W, Ibrahim K. Molecular and Phylogenetic Analysis of Human Papillomavirus Using L1 Gene in Oral Squamous Cell Carcinoma Patients in Baghdad, Iraq. Biomed Pharmacol J 2018;11(2). |
| Copy the following to cite this URL: Al-Malkey M. K, Aldhaher Z. A, Azeez R. A, Hussein S. I, Ismeeal M. C, Mohammed S. W, Ibrahim K. Molecular and Phylogenetic Analysis of Human Papillomavirus Using L1 Gene in Oral Squamous Cell Carcinoma Patients in Baghdad, Iraq. Biomed Pharmacol J 2018;11(2). Available from: http://biomedpharmajournal.org/?p=20389 |
Introduction
Human papillomavirus (HPV) is a circular double-stranded DNA virus, its viral genome composed of three parts: proteins necessary for viral replication and transcription are encoded at the early (E) region; the structural proteins of the viral capsid (L1 and L2) are encoded at the late (L) region; the elements that regulate the viral deoxyribonucleic acid (DNA) replication and transcription are content of the non-coding region.1
The L1 gene, which encodes the major capsid protein is conserved, which make it particularly suitable for to be used as a family taxonomic criterion.2Establishing a distinct HPV “type” is selected when there is at least 10% difference between the DNA sequence of the L1 open reading frame (ORF) and any other characterized type in the cloned viral genome; meanwhile, “subtypes” are referred to the isolates of the same HPV type when the nucleotide sequences of the L1 ORF differ by 2-10%. When the nucleotide sequences of their L1 ORF differ by less than 2 %, referred as a ‘‘variants.’’3,4 Oral cancer (OC) accounted for 300,000 cases and 45,000 deaths occurred worldwide, of which77% were in the less developed regions.5 In United State, approximately 7% of the population will have an oral or oropharyngeal cancer which is generally caused by HPV infection at any given time.6The estimated of sexually active adults is 65%–100%, which have been exposed to HPV at any anatomic site such as oral, genital, or anal, the exposure rate of oral HPV during a lifetime is unknown.7,8The literature had been firmly established etiological agents of anogenital carcinomas with the high-risk human papillomavirus type 16 (HPV-16) and type 18; the morphological similarities, epitheliotropic homing of HPV and its oncogenic potential encouraged the logical assumption of the association between OC and HPV infection.9
HPV-16 as an independent risk factor in 30%-50% of oral squamous cell carcinoma (OSCC) has been assigned according to International Agency for Research on Cancer (IARC) since 2007.10The most common genotype in almost 90% of the HPV positive oropharyngeal cancers (OPC) is HPV- 16.11
The aim of the current study is to detect the HPV genotypes using the L1 gene obtained from OSCC patients and construct a phylogenetic tree.
Materials and Methods
Patients
The current study was approved and under went to the terms of ethical considerations of the Iraqi Ministry of Health. Thirty-five newly diagnosed patients were histopathologically confirmed with OC by two independent pathologists (24 patients were grade I tumors, 4 patients were grade II tumors, and 7 patients were grade III tumors); these patients were attended to maxillofacial surgery clinic of Ghazi Al-Hariri for Specialized Surgery Hospital in Baghdad, Iraq which was enrolled in this study during the period from April 2014 to Jun 2015. An informed consent was taken from each patient. The inclusion criteria for this study were: a) presence of oral cavity cancer (including oral tongue, the floor of mouth, gingiva, lips, buccal mucosa); b) no previous head and neck cancer; c) no prior oncological therapy. The exclusion criteria were: a) patients with a history of autoimmune disorders or systemic diseases; b) previous tumor resection.
Saliva Samples Processing
Unstimulated whole saliva sample up to 5 mL was taken from each patient, which was collected in a 50 mL centrifuge tube and remains on ice whole time during collection. The samples were centrifuged at 2,500 rpm for 15 min at 4°C to spin down exfoliated cells, the saliva supernatant was discarded. Cell pellets were stored at -80°C until further processing.12
DNA Extraction and PCR Analysis
AccuPrep® Genomic DNA extraction kit (Cat# K-3032, Bioneer, Korea) for viral DNA extraction from 200 μl of saliva samples (cell pellet) was used according to the manufacturer’s instructions. HPV-DNA was amplified by conventional PCR assay using specifically designed primer set Forward 5′-ACTGGAAAGGTGCTTGTACC-3′ and Reverse 5′- ACAGGGTTCACAGCCAACAA-3′, to obtain amplicon size (321bp) to target the conserved region according to GenBank reference for alignment of the partial sequence of HPV-L1 gene (GenBank: JX316023.1) using National Center for Biotechnology Information (NCBI)website via Basic Local Alignment Search Tool (BLAST) as previously demonstrated by Agoston et al,(2010).13The amplification was performed using AccuPower® PCR PreMix Kit (Cat# K-2012, Bioneer, Korea) to prepare master mix according to manufacturer’s instructions as follows: 5 μl of template DNA, 1.5 μl of (10 pmol from both primers), and 12 μl of PCR water. The 20 μl reactions were incubated in Thermocycler (MyGene, Korea). Thermocycler condition is listed in Table 1. As a negative control, all the components of the mixture except the DNA template was used. Amplification products of viral DNA were analyzed in 2% polyacrylamide gel electrophoresis containing ethidium bromide, which was placed in TBE buffer and runs at a constant voltage (100V) for approximately 35min. A UV transilluminator was used to visualize the DNA (VISION Gel Documentation System, Scie-Plas, UK). A digital image was captured for the result documentation.
Table 1: PCR Thermo cycling Condition.
| Step | Temperature | Duration | Cycles |
| Initial denaturation | 95°C | 5 minutes | 1 |
| Denaturation | 95°C | 30 seconds | 30 |
| Annealing | 58°C | 30 seconds | |
| Extension | 72°C | 30 seconds | |
| Final extension | 72°C | 5 minutes | 1 |
Sequencing and Phylogenetic Analysis
The amplification products were purified by EZ-10Spin Column DNA Gel Extraction Kit (Cat#BS353, Biobasic, Canada) following the manufacturer’s instructions. HPV genotyping based on Sanger sequencing PCR fragments using Applied Biosystems DNA sequencing analysis was performed in Bioneer Corporation, Korea. The products were deposited in GenBank database and analyzed via BLAST search (http://blast.ncbi.nlm.nih.gov/)with Popset submission (944543671)under the following accession numbers (KT365828–KT365847). Phylogenetic analysis was performed via the neighbor-joining method using the Molecular Evolutionary Genetics Analysis (MEGA) software version 6.0 to identify the evolutionary relationships among the analyzed sequences and a phylogenetic tree was constructed.14
Protein Structure
To study the protein structures corresponding to the sequenced genes, we used the I-TASSER server, which is an Internet-based software product that enables protein structure and function predictions. I-TASSER allows automated generation of high-quality predictions of the 3D structures and biological functions of protein molecules based on their amino acid sequence.15,16
Results
Patients
In the current study 35 newly diagnosed patients (24 males and 11 females). The major histological subtype was squamous cell carcinoma (SCC). This subtype was seen in 26 out of 35 patients accounting for (74%). Adenocarcinoma accounted for (14%) was seen in 5 out of 35 patients and (12%) were other histological subtypes. Grade I Tumors (69%) (24 out of 35). The most common tumor location was the tongue (40%) (14 out of 35) followed by mouth floor (14%) (5 out of 35) and finally, other tumor locations such as maxilla, cheeks were (46%) (16 out of 35).
PCR and Sequencing Analysis
HPV-1 DNA was detected in 20 out of 35 collected saliva samples representing (57%). These HPV-positive samples produced DNA fragments of 321 bp in length for L1 gene, which was amplified using HPV specific primers for partial L1 (GenBank: JX316023.1) (Figure 1). Direct sequencing results from L1 amplicons were analyzed using the BLAST on the NCBI website to confirm HPV-18, HPV-16, and HPV-11 homology. PCR and sequencing analysis revealed that 75% (15 of 20 HPV-positive DNA) were HPV-18 followed by HPV-16 recorded 20% (4 out of 20) and finally 5% (only one out of 20 HPV-positive DNA) was HPV-11.
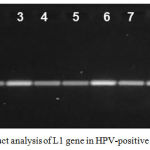 |
Figure 1: PCR product analysis of L1 gene in HPV-positive samples.
|
Where M: marker (2000-100bp), lane (1-10) positive L1 gene at (321bp) PCR product.
Phylogenetic Analysis
A phylogenetic tree was generated using retrieved genome sequences that were deposited under accession number (KT365828–KT365847) and analyzed against 7high risk and low risk HPV genotype references sequences (HPV type18: KF225496.1; HPV type 16: AF140365.1; HPV type 11: AF217526.1;HPV type 33: GQ4790171.1; HPV type 40: HE793060.1; HPV type 58: HM6397161; HPV type 5: AM922325.1); the local HPV isolates in this study were referred as HPV-1, HPV-2, ……, and HPV-20). HPV-18 was the common genotype identified (15 out of 20 samples) representing 75%. The aligned sequences showed high homology to the nucleotide sequence of HPV type 18 (accession numberKF225496.1) (Figure 2). HPV-16 was identified in 4 samples representing (20%), the aligned sequences showed high homology to the nucleotide sequence of HPV type 16 (accession numberAF140365.1) (Figure 3). HPV-11 was identified in only one sample, the aligned sequences showed high homology to the nucleotide sequence of HPV type 11 (accession numberAF217526.1) (Figure 4), the detailed of human papillomavirus local isolates homology to the nucleotide sequence of references from GenBank database are shown in Table 2.
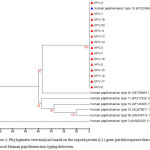 |
Figure 2: Phylogenetic tree analysis based on the capsid protein (L1) gene partial sequence that used for local Human papillomavirus typing detection.
|
The phylogenetic tree was constructed using Unweighted Pair Group method with Arithmetic Mean (UPGMA tree) in (MEGA 6.0 version). The local Human papillomavirus isolates (HPV1, HPV2, HPV3, HPV5, HPV6, HPV7, HPV11, HPV12, HPV13, HPV14, HPV15, HPV17, HPV18,HPV19, and HPV20) were show closed related to NCBI-Blast Human papillomavirus type 18 (KF225496.1), and whereas other NCBI-Blast Human papillomavirus were show out off tree.
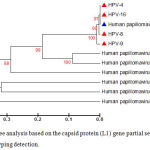 |
Figure 3: Phylogenetic tree analysis based on the capsid protein (L1) gene partial sequence that used for local Human papillomavirus typing detection.
|
The phylogenetic tree was constructed using Unweighted Pair Group method with Arithmetic Mean (UPGMA tree) in (MEGA 6.0 version). The local Human papillomavirus isolates (HPV4, HPV8, HPV9, and HPV16) were show closed related to NCBI-Blast Human papilloma virus type 16 (AF140365.1), and whereas other NCBI-Blast Human papillomavirus were show out off tree.
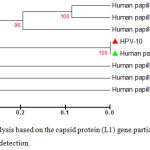 |
Figure 4: Phylogenetic tree analysis based on the capsid protein (L1) gene partial sequence that used for local Human papilloma virus typing detection.
|
The phylogenetic tree was constructed using Unweighted Pair Group method with Arithmetic Mean (UPGMA tree) in (MEGA 6.0 version). The local Human papillomavirus isolates (HPV10) were show closed related to NCBI-Blast Human papillomavirus type 11 (AF217526.1),whereas other NCBI-Blast Human papillomavirus were show out off tree.
The NCBI –BLAST genotypes identity with this study local isolates are revealed in Table 2. The HPV-18 identity match was found with HPV-18 L1 sequence (accession number KF225496.1) isolated from an Indonesian patient infected with cervical cancer. HPV-16 identity match was found with HPV-16 L1 sequence (accession number AF140365.1) isolated from a Chinese patient infected with Condyloma acuminatum. The HPV-11 identity match was found with HPV-11 L1 sequence (accession number AF217526.1) isolated from Chinese patients.
Table 2: Representative genotyping analysis of Human papillomavirus sample isolates based capsid protein (L1) gene according to phylogenetic tree and NCBI –BLAST Genotypes Identity (%) analysis
| No | Local HPV isolate No. | Accession number
(deposited to GenBank) |
NCBI – BLAST Genotypes Identity (%) | Identification HPV Genotype (current study) | ||
| Genotype18
KF225496.1 |
Genotype16
AF140365.1 |
Genotype11
AF217526.1 |
||||
| 1 | HPV-1 | KT365828.1 | 99% | – | – | Genotype 18 |
| 2 | HPV-2 | KT365829.1 | 100% | – | – | Genotype 18 |
| 3 | HPV-3 | KT365830.1 | 100% | – | – | Genotype 18 |
| 4 | HPV-4 | KT365831.1 | – | 100% | – | Genotype 16 |
| 5 | HPV-5 | KT365832.1 | 99% | – | – | Genotype 18 |
| 6 | HPV-6 | KT365833.1 | 100% | – | – | Genotype 18 |
| 7 | HPV-7 | KT365834.1 | 100% | – | – | Genotype 18 |
| 8 | HPV-8 | KT365835.1 | – | 99% | – | Genotype 16 |
| 9 | HPV-9 | KT365836.1 | – | 98% | – | Genotype 16 |
| 10 | HPV-10 | KT365837.1 | – | – | 100% | Genotype 11 |
| 11 | HPV-11 | KT365838.1 | 99% | – | – | Genotype 18 |
| 12 | HPV-12 | KT365839.1 | 100% | – | – | Genotype 18 |
| 13 | HPV-13 | KT365840.1 | 100% | – | – | Genotype 18 |
| 14 | HPV-14 | KT365841.1 | 100% | – | – | Genotype 18 |
| 15 | HPV-15 | KT365842.1 | 100% | – | – | Genotype 18 |
| 16 | HPV-16 | KT365843.1 | – | 98% | – | Genotype 16 |
| 17 | HPV-17 | KT365844.1 | 100% | – | – | Genotype 18 |
| 18 | HPV-18 | KT365845.1 | 100% | – | – | Genotype 18 |
| 19 | HPV-19 | KT365846.1 | 100% | – | – | Genotype18 |
| 20 | HPV-20 | KT365847.1 | 100% | – | – | Genotype 18 |
Protein structure
The protein structure of the L1 gene that isolated from Iraqi HPV-18, HPV-16 was analyzed using the I-TASSER server based on their amino acid sequences as shown in Figure 5 (a, b). Based on this analysis, a ligand-binding site was predicted on L1 that facilitates its binding to heparan sulfate proteoglycan (HSPG) as shown in Figure 5 (c, d).
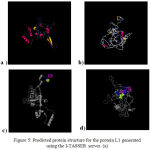 |
Figure 5: Predicted protein structure for the protein L1 generated using the I-TASSER server. (a)
|
The predicted L1 protein structure for HPV-18. (b) The predicted L1 protein structure for HPV-16. (c) A predicted ligand-binding site on L1 for HPV-18. (d) A predicted ligand-binding site on L1 for HPV-16, this site is a possible location for heparan sulfate proteoglycan (HSPG) interactions which crucial for the L1 infectious entry.
Discussion
According to Iraqi cancer registry,17 approximately 4.5% of all cancer cases are oral cancer and about 91.5% of OC are OSCC. In this study, SCC is the most common histological malignant type found in the mouth which agrees with results by (Museedi and Younis, 2014).18 which reported that 91% of OC were SCC; also agrees with earlier reports.19,20–27 Nevertheless, disagrees with Al-Kawaz, (2010) which in his study on the prevalence of OC during 2003-2006 in Iraqi governorates in relation to gender, age, and site found that the OSCC was (41.08%) in Baghdad [28]. The findings of the current study for cancer staging have disagreed with a recent study by Le Campion et al, (2017) in a Brazilian cohort which, reported that (85.1%) were in an advanced stage.29 Unfortunately, a high percentage of oral cancers are only discovered when they become symptomatic, at which point they may be at an advanced stage.21,30–34 Tongue was the primary tumor location in this study which, agrees with Al-Kawaz, (2010) [28], the highest prevalence of tumor location was observed in tongue (55.81%), while the lowest (3.87%) was found in the floor of the mouth also agree with a study by Al Jaber et al, (2016).35 The primary location of OC is an important prognostic factor because the affected anatomic area can determine the accessibility and extension of surgery.36
The papillomavirus major capsid protein, L1, is a ~55 kD protein with the ability to spontaneously self-assemble into virus-like particles (VLPs) which, are potent immunogens, likely due to innate B-cell recognition of the regular icosahedrally displayed spacing of surface epitopes (Bachmann et al., 1993; von Bubnoff, 2012).37–38 The conserved and structural features of L1 make it a good model for viral taxonomy. To the best of our knowledge, the current study is the first in Iraq concerning detection of HPV from saliva exfoliate cells by identification of viral DNA with sets of primers based on HPV partial capsid protein (L1) gene sequence, then genotyping was performed using direct sequencing method.
The phylogenetic analysis of the current study shows that most of the isolates were HPV-18 representing (75%), a 100% identity match was found with an HPV-18 sequence isolated from an Indonesian patient with (accession number KF225496.1), followed by HPV-16 representing (20%) from a Chinese patients with (accession numberAF140365.1).The results of current study disagree with an Indian study by Kabekkodu et al, (2015) and with a study in Argentina by Chiesa et al, (2016), in that HPV-16wasthe most common genotype.39–40 Only a few studies indicated that HPV-18 was the commonest type as in Iranian study by Kermani et al, (2012).41
Papillomavirus capsid initial interaction with the host is largely attributable to L1 interactions with heparan sulfate (HS) carbohydrates displayed on proteoglycans, which is supported by virion binding inhibition and infectious entry by soluble heparin (a highly sulfated form of HS) or by enzymatic removal of HS with heparinase both in vivo, utilizing a murine vaginal challenge model as well as in vitro with cultured cell lines.42–43 In vivo, the heparan sulfate proteoglycan (HSPG) interaction occurs on the extracellular basement membrane, whereas in vitro HPV interact with HSPG on the cell surface and, then to the extracellular matrix (ECM) at a lesser degree.44 Complete dependence upon HSPG binding was found in vivo studies.43 As a result, it appears that all papillomaviruses require initial attachment to HSPGs for successful infection in vivo.45
Conclusion
The genomic characterization of HPV genotypes is pivotal for a deeper understanding of the biological differences of these viruses. There are certain molecular profiles of HPV-positive OSCCs that differentiate them from HPV-negative cancers, as a result, they could be considered as a biological entity separated from the rest of head and neck cancers, and of likely sexual transmission. Unlike most published literature, the highest frequent HPV genotype was HPV-18, which may reflect an intrinsic geographical relatedness and biological differences of HPV-18/HPV-16 and contributes further to research on their infectivity, pathogenicity, and vaccine strategy.
Acknowledgment
We would like to express the sincere gratitude to all seniors of Maxillofacial Surgery Clinic of Ghazi Al-Hariri Hospital for their help, the thanks are extended to all patients who donate their samples in spite of the affliction.
Conflict of Interest
There is no conflict of interest.
Funding Source
There is no funding source.
References
- Betiol J., Villa L. L., Sichero L. Impact of HPV infection on the development of head and neck cancer. Braz J Med Biol Res. 2013;46:217.
CrossRef - de Villiers E. M., Fauquet C., Broker T. R, Bernard H. U., Hausen z. H. Classification of papilloma viruses. Virology. 2004;324:17–27. [PubMed: 15183049].
- Bernard H.U., Burk R. D., Chen Z., Doorslaer v. K., Hausen H., de Villiers E. M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. [PubMed: 20206957].
- Woods R. S. R., O’Regan E. M., Kennedy S., et al. Role of human papillomavirus in oropharyngeal squamous cell carcinoma: a review. World J Clin Cases. 2014;2:172.
CrossRef - Ferlay J., Soerjomataram I., Dikshit R., et al. Cancer incidence and mortality worldwide sources methods and major patterns in GLOBOCAN 2012. Inter J Canc. 2015;136(5):E359–E386. doi:10.1002/ijc.29210. [PMID:25220842].
CrossRef - Pickard R. K., Xiao W., Broutian T. R., et al. The prevalence and incidence of oral human papillomavirus infection among young men and women, aged 18-30 years. Sex Transm Dis. 2012;39:559–66. [PubMed: 22706220].
CrossRef - Hariri S., Unger E. R., Sternberg M., Dunne E. F., Swan D.` Patel S., Markowitz L. E. Prevalence of genital human papillomavirus among females in the United States the National Health And Nutrition Examination Survey, 2003–2006. J Infect Dis. 2011;204:566–73.
CrossRef - Sturgis E. M., Ang K. K. The epidemic of HPV-associated oropharyngeal cancer is here: Is it time to change our treatment paradigms? J Natl Compr Cancer Netw. 2011;9:665–73.
CrossRef - Gonzalez-Ramirez I., Irigoyen-Camacho M., Ramirez-Amador V., et al. Association between age and high-risk human papilloma virus in Mexican oral cancer patients. Oral Dis. 2013;19(8):796–
CrossRef - IARC Monographs on the evaluation of carcinogenic risks to humans. World Health Organization and International Agency for Research on Cancer. Human Papillomaviruses, Lyon. 2007;90.
- Lajer C. B, Buchwald v. C. The role of human papillomavirus in head and neck cancer. Apmis. 2010;118:510.
CrossRef - Michael A., Bajracharya S. D., Yuen P. S., et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–8.
CrossRef - Agoston E. S Robinson S. J Mehra K. K., et al. Polymerase chain reaction detection of HPV in squamous carcinoma of the oropharynx. Am J Clin Pathol. 2010;134:36–41.
CrossRef - Tamura K., Stecher G., Peterson D., Filipski A and Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molec Biolo and Evol. 2013;30(12):2725–29.
CrossRef - Yang J., Yan R., Roy A., Xu D., Poisson J and Zhang Y. The I-TASSER suite protein structure and function prediction .Natur Metho. 2015;12(1):7–8.
CrossRef - Zhang Y. I-TASSER server for protein 3D structure prediction BMC Bioinf. 2008;9:40. doi:10.1186/1471-2105-9-40.
CrossRef - World Health. Organization – Non communicable Diseases (NCD) Country Profiles. 2014.
- Museedi O. S., Younis W. H. Oral cancer trends in Iraq from 2000 to 2008. J Dentl Scie 2014;5:41.
- Chaturvedi A. K., Anderson W. F., Lortet-Tieulent J., et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31(36):4550–59.
CrossRef - Abdul-Hamid G., Saeed N. M., Al-Kahiry W., et al. Pattern of head and neck cancer in Yemen. Gulf J Oncol. 2010;(7):21–24.
- Al-Rawi N. H and Talabani N. G. Squamous cell carcinoma of the oral cavity a case series analysis of clinical presentation and histological grading of 1,425 cases from Iraq. Clin Oral Invest. 2008;12(1):15–18.
CrossRef - Kahan E., Ibrahim A. S., El Najjar K., et al. Cancer patterns in the Middle East – special report from the Middle East Cancer Society. Acta oncol. 1997;36(6):631–36.
CrossRef - Maleki D., Ghojazadeh M., Mahmoudi S. S., et al. Epidemiology of oral cancer in Iran a systematic review. Asian Pac .J Cancer Prev. 2015;16(13):5427–32.
CrossRef - Zini A., Czerninski R and Sgan-Cohen H. D. Oral cancer over four decades epidemiology trends histology and survival by anatomical sites. J Oral Pathol Med. 2010;39(4):299–305.
CrossRef - Subhashraj K., Orafi M., Nair K. V., et al. Primary malignant tumors of orofacial region at Benghazi, Libya: a 17 years review. Cancer Epidemiol. 2009;33(5):332–36.
CrossRef - Chi A. C., Day T. A and Neville B. W. Oral cavity and oropharyngeal squamous cell carcinoma – an update. CA. Cancer. J Clin. 2015;65(5):401–21.
CrossRef - Argiris A., Karamouzis M. V., Raben D., et al. Head and neck cancer. Lancet. 2008;371(9625):1695–1709.
CrossRef - Campion L. A. C. O. V., Ribeiro C. M. B., Luiz R. R., et al. Low Survival Rates of Oral and Oropharyngeal Squamous Cell Carcinoma. Intern J Dentis. 2 017;2017:7. Article ID 5815493, org/10.1155/2017/5815493.
- Maleki D., Ghojazadeh M., Mahmoudi S. S., et al. Epidemiology of oral cancer in Iran a systematic review. Asian Pac J Cancer Prev. 2015;16(13):5427–32.
CrossRef - Halboub E., Abdulhuq M., Al-Mandili A. Oral and pharyngeal cancers in Yemen a retrospective study. East Mediter Health J. 2012;18:985.
CrossRef - Anis R and Gaballah K. Oral cancer in the UAE a multicenter retrospective study. Libyan J Med. 2013;8. doi.10.3402/ljm.v8i0.21782.
CrossRef - Subhashraj K., Orafi M., Nair K. V., et al. Primary malignant tumors of orofacial region at Benghazi, Libya: a 17 years review. Cancer Epidemiol. 2009;33(5):332–36.
CrossRef - Ibrahim N., Al Ashakar M., Gad Z., Warda M., Ghanem H. An epidemiological study on survival of oropharyngeal cancer cases in Alexandria, Egypt. East Mediterr Health J. 2009;15:369–77.
CrossRef - Al-Jaber A., Al-Nasser L and El-Metwally A. Epidemiology of oral cancer in Arab countries. Saudi Med J. 2016;37(3):249– doi: 10.15537/smj.2016.3.11388.
CrossRef - Almeida F. C. S., Cazal C. A., Nunes F. D., Araujo M. E., et al. Prognostic factors in oral cancer Rev Bras Cienc Saude. 2011;15(4):471–78.
- Bachmann M. F., Rohrer U. H., Kundig T. M., et al. The influence of antigen organization on B cell responsiveness. Science. 1993;262:1448–51. [PubMed: 8248784].
CrossRef - Bubnoff v. A. Taking the Gritty Approach. IAVI Report. 2012;16:4–9.
- Kabekkodu S. P., Bhat S., Pandey D., et al. Prevalence of Human Papilloma virus Types and Phylogenetic Analysis of HPV-16 L1 Variants from Southern India. Asian Pac J Cancer Pre. 2015;16(5):2073–80.
CrossRef - Chiesa I. J., Perez M. S., Nunez G. G and Pirola D. A. Genetic variability and phylogeny analysis of partial L1 gene of human papillomavirus variants in Buenos Aires, Argentina. Virus Dis. 2016;27(1):41–47. doi 10.1007/s13337-015-0295-3.
CrossRef - Kermani A. I., Seifi S. H., Dolatkhah R., et al. Human Papilloma Virus in Head and Neck Sequamous Cell Carcinoma. Iran J Cancer Prev. 2012;1:21–26.
- Giroglou T., Florin L., Schafer F., et al. Human papilloma virus infection requires cell surface heparan sulfate. J Virol. 2001;75:1565–70. [PubMed: 11152531].
CrossRef - Johnson K. M., Kines R. C., Roberts J. N., et al. Role of heparan sulfate in attachment to and infection of the murine female genital tract by human papilloma virus. J Virol. 2009;83:2067–74. [PubMed: 19073722].
CrossRef - Selinka H. C., Florin L., Patel H. D., et al. Inhibition of transfer to secondary receptors by heparan sulfate-binding drug or antibody induces noninfectious uptake of human papilloma virus. J Virol. 2007;81(109):70–80. [PubMed: 17686860].
CrossRef - Buck C. B., Day P. M and Trus B. L. The papillomavirus major capsid protein L1. Virology. 2013;445(0):169–74. doi: 10.1016/j.virol.2013.05.038. Epub 2013 Jun 22.
CrossRef







