Satya Prakash Sahu1 , Priyanka Agrawal1
, Priyanka Agrawal1 , Narendra D Londhe2
, Narendra D Londhe2 and ShrishVerma3
and ShrishVerma3
1Departmentof Information Technology, National Institute of Technology, Raipur, India;
2Departmentof Electrical Engineering, National Institute of Technology, Raipur, India;
3Departmentof Electronics and Telecommunication Engineering, National Institute of Technology, Raipur, India;
Corresponding Author E-mail: spsahu.it@nitrr.ac.in.
DOI : https://dx.doi.org/10.13005/bpj/1315
Abstract
For computer-aided-diagnosis (CAD) System, the lung segmentation phase is having most significant role in the detection of lung cancer at initial stages. It is needed as preprocessing step for obtaining the accurate Region of Interest (ROI) area. Efficiency of CAD system is mainly depending on how the lungs are precisely segmented. The effective lung segmentation overcomes the various challenges offered in CAD system to deal with the cases of juxtapleural nodules. This paper emphasizes the proposed method for lung segmentation in CT images using clustering approach of fuzzy-c-means with automatic thresholding and morphological operations. The experimental database contains 20 patients’ series (approximately 3600 images) from publically available LIDC-IDRI dataset which includes 10 general cases and 10 cases having juxtapleural nodules in pulmonary region. Reference standard contours are prepared by the expert through manually tracing the lungs boundary. The segmented lungs obtained through proposed method are compared with the reference standards with the use of various parameters. The proposed method achieved overall overlap ratio of 99.94% accuracy with 0.94 Jaccard’s Index and 0.97 Dice similarity coefficient values
Keywords
CAD System; Computed Tomography;Fuzzy c-means;Lung Segmentation; Thresholding; Morphological Operations Pulmonary nodules;
Download this article as:| Copy the following to cite this article: Sahu S. P, Agrawal P, Londhe N. D, Verma S. A New Hybrid Approach Using Fuzzy Clustering and Morphological Operations for Lung Segmentation in Thoracic CT Images. Biomed Pharmacol J 2017;10(4). |
| Copy the following to cite this URL: Sahu S. P, Agrawal P, Londhe N. D, Verma S. A New Hybrid Approach Using Fuzzy Clustering and Morphological Operations for Lung Segmentation in Thoracic CT Images. Biomed Pharmacol J 2017;10(4). Available from: http://biomedpharmajournal.org/?p=17589 |
Introduction
As per various statistics surveyed by different agencies the lung cancer has been observed as most common cancer worldwide.In 2012, 1.8 million new lung cancer cases were estimated.1 Deaths due to lung cancer are more as compared to other cancer related death worldwide. It contributes 13% for the cases registered as cases and in the field of deaths due to cancer its contribution was 19% in 2012, reported by GLOBOCAN 2012 (IARC) Section
![Figure 1: Worldwide Incidence and Mortality rate [Courtesy of GLOBOCAN 2012 (IARC) Section of Cancer Surveillance] of Cancer Surveillance (Fig 1) [1].](http://biomedpharmajournal.org/wp-content/uploads/2017/11/Vol10No4_New_Suj_fig1-150x150.jpg) |
Figure 1: Worldwide Incidence and Mortality rate [Courtesy of GLOBOCAN 2012 (IARC) Section of Cancer Surveillance] of Cancer Surveillance (Fig 1) [1].
|
In US, 224,400 cases are new cases of cancer which is about 14% and 158,080 cases related to cancer death are 27% as reported by American Cancer Society, Cancer facts and figure 2016.2 In India, its percentage was 6.9% for all new cancer cases and 9.3% for cancer related deaths. This data includes both male and female patients. It has been observed that most of the cancer cases are registered from Mizoram for both male and female (Age adjusted rate 28.3 and 28.7 per 100,000 population in males and females, respectively).3
Lung cancer leading to mortality rate higher in subsequent years and this encourages many researchers for the diagnosis of lung cancer in early stages. The survival rate may be increased by 70-80%, if this cancer is detected and diagnosed in prior stages.4 The first screening test using thorax X-ray modality, the large nodule can only be detected, for small nodule of size even upto 2 cm could not be detected since of their unfavorable positions and locations.5 With the advent of multi-slice, spiral CT, capability to perform a 3D reconstruction of the anatomical structure of human body, has explored detection of lung cancer in the early stages6 and small nodules with a diameter lower than 5 mm (micro-nodules) can still be caught by LD-CT.
In a typical computer-aided diagnosis (CAD) system for lung cancer,7,8 the lung segmentation is very important step for the detection of lung cancer or abnormalities and pulmonary diseases in lung area. For detecting proper lung region, CAD system have to overcome from various challenges like inhomogeneities in lung region and similar densities in other pulmonary structures like veins, bronchi, arteries, and bronchioles. Among all these challenges, the most important and difficult task is to determine the nodules attached to lung wall (i.e. juxtaplueral). Chances of missing the juxtaplueral nodules get increased if the lungs are not segmented properly. Approximately 5% to 17% true lung nodules are not detected because of poor and inaccurate lung segmentation, as stated by Armato et al.9 For the above stated reason it is necessary to effectively and accurately segment the lungs so that juxtaplueral nodules get properly detected.
In this paper, FCM algorithm is deployed for determining lung contour and to extract the pulmonary field or ROI for further detection of various types of nodules in these regions. The segmentation follows the correction of broken contour based on various morphological operations with the inclusion of juxtaplueralnodules, this phase also reduces the scope of over-segmentation. For testing the proposed method, approximately 3600 CT slices from 20 cases of LIDC-IDRI (Lung Imaging Database Consortium & Image Database Resource Initiative) dataset have been taken.10,11 The experts have been asked for trace the contour of lungs to create the reference images of CT slices. The obtained results have been compared and evaluated with the various performance metrics such as Jaccard’s similarity index, Dice similarity coefficient, over-segmentation rate, under-segmentation rate, and overlap-ratio difference. This paper remaining section follows – Section II includes the review of previous work and also these research works are evaluated on the basis of challenges, issues, and effectiveness. In Section III, material and methods are presented in the following steps: data acquisition, preprocessing of data, and lung segmentation by using FCM with thresholding and morphological operations. Section IV describes experiments, evaluations of proposed method and results. In the last Section V, the results of proposed work is compared with some other works as conclusions and which is followed by limitations of the method and discussion of scopes of future work in this domain.
Related Work
For lung, segmentation, various automatic and semi-automatic approaches have been addressed by the authors. Automation level, processing time and the accuracy terms such as sensitivity & specificity are the measuring parameter for effectiveness and efficiency of the existing methodologies. The studies of these methods of segmentation mainly rely on the criteria like thresholds, shape based, active-contour, region based, clustering and morphological operations. For image segmentation of object the most general method is gray level or intensity value based technique and is commonly known as Threshold based segmentation. The region of Lung lobes are having lower intensity level i.e. approximately -500 HU as compared to surrounding anatomical structure of thorax, due to this reason authors have adopted optimum threshold as their basis for segmenting the lungs in their research studies. Hu et al.12 used iterative automatic threshold with morphological operation for smoothing the images in their irregular boundaries. Wei et al.13 analyses histogram for threshold to segment the pulmonary region. Ye et al.14 had given the Adaptive fuzzy thresholding for lung segmentation in CT images. Helen et al.15 used the particle swarm optimization (PSO) for the optimal 2D-Otsu thresholding algorithm for segmentation of lung parenchyma.
Shape based segmentation is another useful technique. Prior shape information of lungs are registered for the collected dataset and used further for extraction of ROI from the thorax region. This method uses an energy framework to help and guide deformable models in lung lobe segmentation and for such framework the parameters like edges or contour, points, etc. are characterized for previous data. For the local minima issues in lung segmentation Annangi et al.16 proposed region based active contour technique for x-ray images by using initial shape and low level features. Kockelkorn et al.17 took the severe abnormal data with their prior shape parameters and trained using k-NN classifier and devised interactive techniques for lung. Pattern recognition technique with the combination of statistical shape model and anatomical information for the robust lung segmentation has been adopted by Sofka et al.18 Pu et al.19 proposed “break-and-repair” technique for segmentation of a medical image based on geometric modeling and shape. An approach given by Sun et al.20 based on active shape model for the segmentation of lungs boundary and further to adapt a method based on constrained optimal surface using initial result of segmentation. Gill et al.21 gave the feature based atlas approach for the initialization of active shape models in CT images.
The other significant method used in various articles22-25 of lung segmentation is based on Active contour method (ACM). The main fundamental concept behind this method is minimizing energy function iteratively through dynamic contour at each step. The most challenging part of this approach is the problem of initialization and convergence. Chenyang Xu and Jerry L Prince22 proposed Gradient Vector Flow (GVF) as an external force for guiding the snake so that better convergence can be obtained. Wang et al.23 further work for improving the initialization and convergence by Normally Biased Gradient Vector Flow (NBGVF). Cui et al.24 manipulate the ACM based on statistics of local region for medical images segmentation. An automatic method using ACM is exploited fully by Athertya et al.25 that helps in reducing the user interaction for the segmentation of lungs.
Recently, one state-of-art method proposed by Soliman et al.38 for accurate lung segmentation based on guided shape modeling. This approach provides the stack of CT images as 3D joint Markov-Gibbs random field (MGRF) model. The method segment the pathological lungs due to integrating the first and second order probabilistic descriptors of original and Gaussian scale space filtered image with prior guided adaptive shapes. However, this method requires larger training set of healthy and pathological lungs as guided landmarks for better performance.
The existing above approaches are based on general lung segmentation techniques. These methods are either not considering the cases of CT scan having the juxtapleural nodules or not so specific to deal with the complexity arises due to the presence of such nodules attached with the parenchyma of lungs. In this paper we are concerned about this complexity and prepared our database by including the 50% data with such cases and developed the framework to segment the accurate lung region even in the cases with CT images having juxtapleural nodules.
Material and Methods
Data
The database for the experiment contains 20 subjects and has been prepared from publically available LIDC-IDRI data set.10 The database of LIDC-IDRI has 1018 cases of Lung CT scans under The Cancer Imaging Archive (TCIA) Public Access.26
The collection of lung cancer thoracic CTs with marked-up annotation introduced by National Cancer Institute (NCI), later on upgraded by Foundation for the National Institutes of Health (FNIH) and Food and Drug Administration (FDA) under public-private partnership for the evaluation, training and development of CAD systems and specifies process for the detection and diagnosis of lung cancer. The annotation of this database has been done by four experts (radiologists) and stored in XML file.11 The database stored all Lung CT images with specifications as follows: DICOM image of size 512 X 512 pixels, tube current in the range of 265 to 570 mAs, tube voltage 120kVp and intensity level from -600 to 1600 HU.11
Data Preprocessing
Data preprocessing is required for making the selected data as suitable for computable format. It starts with selecting the suitable cases from LIDC-IDRI database. The steps for the data preprocessing are shown in Fig. 2. In this experiment, 20 patient CT scan images have been taken with 10 cases of juxtapleural lung nodules. The contour or boundary tracing of lung region for each patient of all the slices are done by expert as the representation of reference standard or ground truth. The accuracy can be further enhanced if the image pixel value is converted to double format. The optimum level of contrast for the selected data is set by the process of image enhancement.The acquired images of LIDC possess DICOM format data so it need to be converted into Hounsfield Unit (HU) and to set window size (level and width) appropriately for the segmentation of lungs.
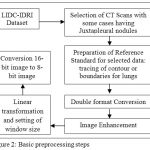 |
Figure 2: Basic preprocessing steps
|
Linear transformation performs the rescaling of slope and intercept and lead to defined range of HU values of the images. The final step for getting preprocessing is the conversion from 16-bits images into 8-bits images so as to get the suitable data for further process.
Lung Segmentation
The proposed method for lung segmentation is shown in Fig. 3 as flowchart and is implemented by the following steps:
Applying Fuzzy-c-means clustering based segmentation to generate the gray-scale masked image of the CT slices. The derivation & steps of the algorithm are given in section 3.3.1.
By using automatic thresholdingor otsu’s algorithm, the output of fuzzy-c-means mask is converted into binary image (method discussed in section 3.3.2) followed by the background subtraction for ROI in image.
Large airways, blood vessels and trachea are removed with the application of hole-filling This algorithm is based on the process of dilation, intersection and complementation.
Let I be the set whose boundary pixel are labeled by 1s (considering 8-connected boundaries) & each boundary surrounding a hole (background region). With the given point x in each hole, the goal is to fill the holes by 1s. This step is followed by morphological reconstruction for removing the lighter border by reducing the overall intensity of the border structures.
The morphological closing operations are applied to deal with the cases having juxtapleural nodules.
At last masking the output image with input gray-scale image for obtaining the segmented lung region.
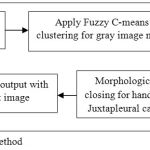 |
Figure 3: Flowchart for the proposed method
|
Fuzzy-c-means Algorithm
Fuzzy c-means (FCM) is a method of clustering and is used here for lung segmentation. The flexible sense of this method is that a data point can belong to two or more cluster at a time. FCM is initially given by Dunn27 and further enhanced by Bezdek.28 The algorithm is based on minimization of the objective function (Jp)
![]()
where, p is any real number greater than 1 i.e.1 ≤ p < ∞, vij is the membership matrix of xi in the cluster j, xi is ith of d-dimensional data measured (say pixel intensity), ci is the centroid or cluster center and ||*|| is a norm representing the similarity between any considered pixeland thecluster center.
Based on the optimization of above objective function fuzzy partitioning is performed. The membership vij and the centroid ci are updated as follows:
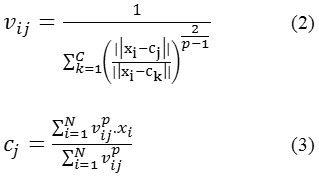
The iteration will terminate
when
![]()
where Φ is a stopping criterion ranges 0<Φ<1 and k as the iteration steps. This procedure follows the converging toward a local minimum of jp.
The step wise algorithm follows as:
Image intensity distribution computed for different gray-scale value.
Initialize V=[vij ] matrix, V(0)
At kth step: calculate the centroid vectors C(k)=[ci ] with V(k)as per eq. (3).
Update membership matrices V(k), V(k+1)as per eq. (2).
For termination, if ||V(k+1) – V(k)||<Φ then STOP; else return to step (c).
Automatic Threshold Algorithm
Otsu’s 29 had given the automatic method of thresholding. The estimated threshod of gray level value (g*) is obtained by simple and effective method that minimizes the weight within-class variance and maximizes the between class variance assuming that the histogram are bimodal. For the given image, the representation of pixels in M gray levels [1, 2…, M], the normalized histogram as probability distribution ( ) can be given as:
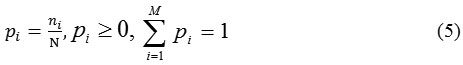
where, represents the number of pixels at the gray level i, and N= n1 +n2 +…+nm is thetotal number of pixel.
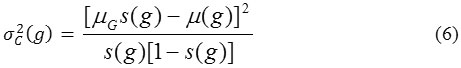
where, s(g) and μ (t) / s (t) represent class occurrence probability and gray level mean value respectively of background,[1- s(g)] and μG – μ(g)) / (1 – s(g)) represent class occurrence probability and gray level mean value respectively of object.
Thus with the analysis of histogram and checking for each gray level, the general form of optimal threshold g* that maximize
![]()
can be defined as:
![]()
Result Analysis
Experimental database contains 20 patients or CT series (approximately 3600 slices or images examined) out of which 10 patients having juxtaplueral nodules in their CT images, taken from the LIDC-IDRI dataset. The lung boundaries or contours were manually traced by experts under the observation or supervision of thoracic radiologists for each case. These tasks have been performed by an open-source system “The Medical Imaging Interaction Toolkit”, a workbench and toolkit of MITK 2016.11.30 This workbench has deployed here as annotation tool that includes Visualization Toolkit (VTK) and Insight Toolkit (ITK). The flexibility with this toolkit is that it support border corrections and contour adjustments if some inaccuracies are observed. These manually traced boundaries or contours were considered as ground truth for respective images. Finally for the evaluation of experimental results of proposed method of lung segmentation with reference standard or ground truth various evaluation measures have been used.
Evaluation Measures
The following metrics have been utilized for the performance of results of lung segmentation:over-segmentation & under-segmentation rate, overlap ratio, Dice similarity coefficient andJaccard’s similarity index.
The over-segmentation is given as the area (number of pixel) or volume (number of voxels) that is included as a portion of the segmented ROI (result of proposed method) but are absent in reference standard or ground truth image whereas under-segmentation is given as the area or volume that are included in ground truth, but are absent in segmented ROI. 31,32 The volume overlap ratio (VO) is given as the relative overlap between two volumes i.e. segmentation mask produced by our proposed method (Vm) and ground truth (Vg) and formulated by taking the intersection Vmand Vg divided by their union.33 The definitions of these measures are as follows:
The over-segmentation rate Os (Vm, Vg ) is given by
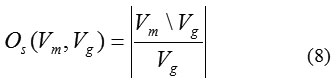
whereVm\Vg is the relative compliment of Vm in Vg. In the similar manner the under-segmentation rate Us (Vm,Vg) is given by
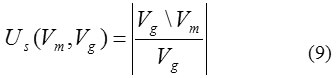
whereVg\Vm is considered as the relative compliment of Vg in Vm. The volume overlap ratio (VOR) can be defined as
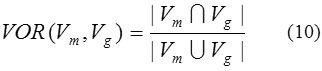
The other measures which have been used for the segmentation accuracies in the terms of similarity between the segmentation results and ground truths are Dice-similarity coefficient (DSC) and Jaccard’s similarity index (JSI).34 These methods compare the similarity between the segmented ROIs (number of pixels or voxels) of adopted method (Rm) to the ground truth region (Rg).
The Jaccard’s similarity index is given as the overlap degree between Rmand Rgand follows as
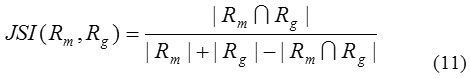
Similarly Dice-similarity coefficient is defined as the overlap of two regions i.e. Rmand Rgand can be given as
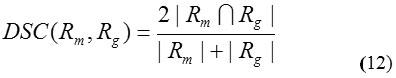
The values of JSI and DSC will be in range of [0,1] and higher values indicates better results in terms of segmentation accuracy.
Results and Discussion
The performance evaluation of our proposed method has been tested on 20 CT scans (approximately 3600 CT slices)
including 10 CT scans of juxtapleural nodules. Fig.4 & Fig.5 show the sample results (randomly selected slice of 5 CT scans each of the two cases) stepwise for our adopted method in lung segmentation for general cases and cases having juxtapleural nodules respectively.
With the use of evaluation measures (section 4.1), in order to show the average distance of segmentation error of 20 patients (each patient having 150-200 slice images) undertaken are represented in horizontal-axis (from 1 to 20) in Fig.6 (a), we depicts bar plots of cumulative probability distribution based on over-segmentation, under-segmentation and overlap ratio difference error in vertical-axis. Similarly in Fig.6 (b) the accuracies are represented in terms of similarity measures i.e. bar plots of the cumulative probability distribution based on the Dice-similarity coefficient and Jaccard’s similarity index (both represented in vertical-axis) for 20 patients (represented in horizontal-axis) are shown.
In the Table 1 we have shown the overall results of our adopted method for the all data that achieved value (mean values) : over-segmentation of 0.0394, under-segmentation of 0.0193, and overlap ratio difference of 0.0587, overlap ratio of 99.9413, Dice-similarity coefficient of 0.9710 and Jaccard’s index of 0.9444. For the cases having juxtapleural nodules our approach achieved an outstanding performance (in mean values) i.e overlap ratio of 99.9347% and Dice-similarity coefficient value of 0.9679 and Jaccard’s index value of 0.9389.
The number of authors had given various methods and techniques for lung segmentation, but only some are effective for the cases of juxtapleural nodules. Table 2 compares the performance for our approach with the other methods for lung segmentation. The proposed method achieved the best average overlap ratio among the others listed in the table. In terms of Jaccard’s similarity index the proposed method performs better than Liao.37 Although, for fair comparison of methods is possible if the same datasets and image standards have accessed. But due to non availability of other database we have taken the public datasets and tested our hybrid approach and hence it is not possible for complete comparison. However comparing to other approaches, our proposed method is achieving the good results and generating the better accuracies for lung segmentation.
Table 1: Statistical analysis of Over-segmentation, Under-segmentation, Overlap ratio difference, Dice similarity coefficient and Jaccard’s similarity index of proposed method.
| Over-segmentation | Under-segmentation | Overlap-ratio diff. | Dice similarity Coefficient | Jaccard similarity Index | |
| Mean | 0.039428 | 0.019319 | 0.058747 | 0.971036 | 0.944359 |
| SD | 0.026018 | 0.017359 | 0.036979 | 0.033143 | 0.017973 |
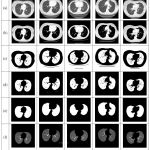 |
Figure 4: The lung segmentation results of our proposed approach for the images of general cases.
|
Row (a) are five original lung CT images (b) gray scale masked images after applying FCM (c) binary images of previous step using Otsu’s threshold (d) background subtracted images; (e) final lung mask after removing the large airways, trachea, and blood vessels and (f) final result– segmented lungs.
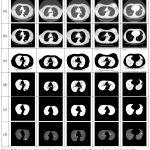 |
Figure 5: The lung segmentation results of our proposed approach for the image having juxtapleural nodules.
|
Row (a) are five original lung CT images (b) gray scale masked images after applying FCM (c) binary images of previous step using Otsu’s threshold (d) background subtracted images; (e) final lung mask after removing the large airways, trachea, and blood vessels and (f) final result– segmented lungs.
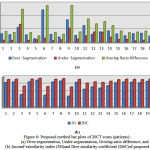 |
Figure 6: Proposed method bar plots of 20CT scans (patients): (a) Over-segmentation, Under-segmentation, Overlap ratio difference, and (b) Jaccard’ssimilarity index (JSI)and Dice similarity coefficient (DSC)of proposed method.
|
Table 2: Performance comparison of proposed method for segmentation of lungs.
| Authors &
Works |
Datasets | Overlap-ratio | Over Segment- ation | Under Segmenta-tion | Dice Similarity Coefficient | Jaccard Similarity Index |
| Wei et al. [35] | 97 nodules CT scans25 juxtapleural nodule scans | 95.24% | – | – | – | – |
| S.Zhou et al.[36] | 20 CT scans with 65juxtapleural nodules | 95.81% | – | – | – | – |
| Liao et al. [37] | 80 cases divided in 4 categories of non nodules, benign nodules, malignant SPN and pleural nodules with 20 cases in each category | 92.22% | – | – | – | Non nodules 0.941
Benign nodules 0.9342 Malignant SPN 0.9257 Pleural nodules 0.8821 |
| Proposed method | 20 cases of LIDC-IDRI datasets with 10 cases of juxtapleural nodules (3600 CT images approx.) | 99.94% | 0.0394 | 0.0193 | 0.9710 | Overall 0.9444
Pleural nodules 0.9389 |
Conclusion
Lung segmentation is very crucial and important step in CAD system for the detection and diagnosis of lung cancer in prior stages. For the past few decades’ number of research works have been given by various authors for lung segmentation and shown their effectiveness in different cases. In this paper, we have implemented a new hybrid approach using fuzzy clustering (FCM) with morphological operations for the segmentation of lungs in CT images that have shown the very effective results in the cases of lungs having juxtapleural nodules.
The efficiency of our approach is tested on 20 cases (i.e. 10 general and 10 cases of juxtapleural nodules) of lung thoracic CT scans from LIDC-IDRI a public dataset. The experimental results show that the proposed method can correctly including all juxtapleural nodules into the ROI of lungs with the minimum rate of under and over-segmentation and thereby achieved overall 99.94% of overlap ratio. In the result section, we have also given the performance in terms of of Dice similarity coefficient and Jaccard’s index and shown the accuracies of 0.9710 and 0.9444 respectively. The average processing time for the adopted method is 0.80 s per CT slice using MATLAB software experimented on PC with CPU Intel Core i5 3.1 GHz and 8 GB RAM. The proposed approach can fulfill the requirements of CAD system for the lung cancer and provide the accurate search space for further processing toward the detection and classification of pulmonary nodules.
Acknowledgement
The authors would like to thank the Director of National Institute of Technology, Raipur and Dr. S. Pandey, Head, Department of Information Technology for the necessary support to do this study.
Conflict of Interest
No conflict of interest
Funding Source
There was no funding support from any agencies.
References
- Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. France L. International Agency for Research on Cancer; 2013. GLOBOCAN. 2012;(1)0. Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Available from: http://globocan.iarc.fr , accessed on 04/10/2016.
- American Cancer Society, Cancer facts and figures. 2016.
- Indian Council of Medical Research; National Cancer Registry Programme. Three Year Report of Population Based Cancer Registries. 2013;2009-2011. Available from: http://www.ncrpindia.org.
- Swensen S.J, Jett J.R, Sloan J.A, Midthun D.E, Hartman T.E, Sykes A.M, Aughenbaugh G.L, Zink F.E, Hillman S.L, Noetzel G.R, Marks R.S. Screening for lung cancer with low-dose spiral computed tomography. American journal of respiratory and critical care medicine. 2002 Feb 15;165(4):508-13.
CrossRef - Flehinger B.J, Melamed M.R, Zaman M.B, Heelan R.T, Perchick W.B, Martini N. Early Lung Cancer Detection: Results of the Initial (Prevalence) Radiologic and Cytologic Screening in the Memorial Sloan-Kettering Study 1–3. American Review of Respiratory Disease. 1984 Oct;130(4):555-60.
- Awai K, Murao K, Ozawa A, Komi M, Hayakawa H, Hori S, Nishimura Y. Pulmonary Nodules at Chest CT: Effect of Computer-aided Diagnosis on Radiologists’ Detection Performance 1. Radiology. 2004 Feb;230(2):347-52.
CrossRef - El-Baz A, Beache G.M, Gimel’farb G, Suzuki K, Okada K, Elnakib A, Soliman A, Abdollahi B. Computer-aided diagnosis systems for lung cancer: challenges and methodologies. International journal of biomedical imaging. 2013 Jan 29;2013.
- Antonelli M, Cococcioni M, Lazzerini B, Marcelloni F. Computer-aided detection of lung nodules based on decision fusion techniques. Pattern Analysis and Applications. 2011 Aug 1;14(3):295-310.
CrossRef - Armato S.G, Sensakovic W.F. Automated lung segmentation for thoracic CT: Impact on computer-aided diagnosis1. Academic Radiology. 2004 Sep 30;11(9):1011-21.
CrossRef - Armato I.I.I, Samuel G, Geoffrey M , Luc B, McNitt-Gray M.F,Charles M.R, Anthony P.R. Laurence C.P. Data From LIDC-IDRI. The Cancer Imaging Archive. 2015. http://doi.org/101.7937/K9/TCIA.2015.LO9QL9SX
- Armato S.G, McLennan G, Bidaut L, McNitt‐Gray M.F, Meyer C.R, Reeves A.P, Zhao B, Aberle D.R, Henschke C.I, Hoffman E.A, Kazerooni EA. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Medical physics. 2011 Feb 1;38(2):915-31.
CrossRef - Hu S, Hoffman E.A, Reinhardt J.M. Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE transactions on medical imaging. 2001 Jun;20(6):490-8.
CrossRef - Wei Q, Hu Y, Gelfand G, MacGregor J.H. Segmentation of lung lobes in high-resolution isotropic CT images. IEEE Transactions on biomedical engineering. 2009 May;56(5):1383-93.
CrossRef - Ye X, Lin X, Dehmeshki J, Slabaugh G, Beddoe G. Shape-based computer-aided detection of lung nodules in thoracic CT images. IEEE Transactions on Biomedical Engineering. 2009 Jul;56(7):1810-20.
CrossRef - Helen R, Kamaraj N, Selvi K, Raman V.R. Segmentation of pulmonary parenchyma in CT lung images based on 2D Otsu optimized by PSO. InEmerging Trends in Electrical and Computer Technology (ICETECT), 2011 International Conference on. IEEE. 2011 Mar 23;536-541.
- Annangi P, Thiruvenkadam S, Raja A, Xu H, Sun X, Mao L. A region based active contour method for x-ray lung segmentation using prior shape and low level features. InBiomedical Imaging: From Nano to Macro. 2010 IEEE International Symposium on. IEEE. 2010 Apr 14;892-895.
- Kockelkorn T.T, van Rikxoort E.M, Grutters J.C, Ginneken v.B. Interactive lung segmentation in CT scans with severe abnormalities. InBiomedical Imaging: From Nano to Macro, 2010 IEEE International Symposium on . IEEE. 2010 Apr 14;564-567.
CrossRef - Sofka M, Wetzl J, Birkbeck N, Zhang J, Kohlberger T, Kaftan J, Declerck J, Zhou S. Multi-stage learning for robust lung segmentation in challenging CT volumes. Medical image computing and computer-assisted intervention–MICCAI 2011. 2011;667-74.
- Pu J, Paik D.S, Meng X, Roos J, Rubin G.D. Shape “break-and-repair” strategy and its application to automated medical image segmentation. IEEE transactions on visualization and computer graphics. 2011 Jan;17(1):115-24.
CrossRef - Sun S, Bauer C, Beichel R. Automated 3-D segmentation of lungs with lung cancer in CT data using a novel robust active shape model approach. IEEE transactions on medical imaging. 2012 Feb;31(2):449-60.
CrossRef - Gill G, Toews M, Beichel R.R. Robust initialization of active shape models for lung segmentation in CT scans: A feature-based atlas approach. Journal of Biomedical Imaging. 2014 Jan 1;2014:13.
- Xu C, Prince JL. Snakes, shapes, and gradient vector flow. IEEE Transactions on image processing. 1998 Mar;7(3):359-69.
CrossRef - Wang Y, Liu L, Zhang H, Cao Z, Lu S. Image segmentation using active contours with normally biased GVF external force. IEEE Signal Processing Letters. 2010 Oct;17(10):875-8.
CrossRef - Chan T.F, Vese L.A. Active contours without edges. IEEE Transactions on image processing. 2001 Feb;10(2):266-77.
CrossRef - Athertya J.S, Kumar G.S. Automatic initialization for segmentation of medical images based on active contour. InBiomedical Engineering and Sciences (IECBES), 2014 IEEE Conference on. IEEE. 2014 Dec 8;446-451.
CrossRef - Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. Journal of digital imaging. 2013 Dec 1;26(6):1045-57.
CrossRef - Dunn J C. A Fuzzy Relative of the ISODATA Process and Its Use in Detecting Compact Well-Separated Clusters. Journal of Cybernetics. 1973 Sep;3(3):32-57.
CrossRef - James B.C. Pattern recognition with fuzzy objective function algorithms. Springer Science & Business Media. 2013.
- Otsu N. A threshold selection method from gray-level histograms. Automatica. 1975 Jun;11(285-296):23-7.
- MITK: The Medical Imaging Interaction Toolkit, German Cancer Research Center, Division of Medical and Biological Informatics, http://mitk.org/wiki/MITK. 2016.
- Pu J, Roos J, Chin A.Y, Napel S, Rubin G.D, Paik D.S. Adaptive border marching algorithm: automatic lung segmentation on chest CT images. Computerized Medical Imaging and Graphics. 2008 Sep 30;32(6):452-62.
CrossRef - Hoover A, Jean-Baptiste G, Jiang X, Flynn P.J, Bunke H, Goldgof D.B, Bowyer K, Eggert D.W, Fitzgibbon A, Fisher R.B. An experimental comparison of range image segmentation algorithms. IEEE transactions on pattern analysis and machine intelligence. 1996 Jul;18(7):673-89.
CrossRef - van Rikxoort E.M, de Hoop B, Viergever M.A, Prokop M, Ginneken v.B. Automatic lung segmentation from thoracic computed tomography scans using a hybrid approach with error detection. Medical physics. 2009 Jul 1;36(7):2934-47.
CrossRef - Sampat M.P, Wang Z, Gupta S, Bovik A.C, Markey M.K. Complex wavelet structural similarity: A new image similarity index. IEEE transactions on image processing. 2009 Nov;18(11):2385-401.
CrossRef - Wei Y, Shen G, Li J.J. A fully automatic method for lung parenchyma segmentation and repairing. Journal of digital imaging. 2013 Jun 1;26(3):483-95.
CrossRef - Zhou S, Cheng Y, Tamura S. Automated lung segmentation and smoothing techniques for inclusion of juxtapleural nodules and pulmonary vessels on chest CT images. Biomedical Signal Processing and Control. 2014 Sep 30;13:62-70.
CrossRef - Liao X, Zhao J, Jiao C, Lei L, Qiang Y, Cui Q. A Segmentation Method for Lung Parenchyma Image Sequences Based on Superpixels and a Self-Generating Neural Forest. PloS one. 2016 Aug 17;11(8):e0160556.
CrossRef - Soliman A, Khalifa F, Elnakib A, El-Ghar M.A, Dunlap N, Wang B, Gimel’farb G, Keynton R, El-Baz A. Accurate lungs segmentation on CT chest images by adaptive appearance-guided shape modeling. IEEE transactions on medical imaging. 2017 Jan;36(1):263-76.
CrossRef








