Manuscript accepted on :April 14, 2017
Published online on: --
Plagiarism Check: Yes
Jeba Shiney1, O. J. Amar Pratap Singh2 and Priestly Shan. B3
1Noorul Islam University, Kumarakoil, Thuckalay, India.
2Director(Administration), Noorul Islam University, Kumarakoil, Thuckalay, India
3Eranad Knowledge City, Manjeri, Malappuram, Kerala.
Corresponding Author E-mail: jebashiney@gmail.com
DOI : https://dx.doi.org/10.13005/bpj/1266
Abstract
In this paper, various algorithms and techniques, which aids in development of secondary observer systems for Downsyndrome detection in fetus of first and second trimester is being evaluated and presented. 50papers from 1982 to 2016 has been reviewed and the consolidated study is being presented. The parameters for comparison include Markers for Down Syndrome Detection, Algorithms, Sensitivity, Specificity and accuracy of these algorithms. Markers for study include Nasal Bone Length, Nuchal Translucency Thickness, and Naso Frontal Angle. These markers have been utilized by researchers for design of Secondary Observer systems and have achieved various degrees of accuracies.
Keywords
Accuracy; Down Syndrome; Markers; Nasal Bone Length; Nuchal TranslucencySecondary Observer Systems; Specificity; Sensitivity;
Download this article as:| Copy the following to cite this article: Shiney J, Singh O. J. A. P, Shan B. P. A Review on Techniques for Computer Aided Diagnosis of Soft Markers for Detection of Down Syndrome in Ultrasound Fetal Images. Biomed Pharmacol J 2017;10(3). |
| Copy the following to cite this URL: Shiney J, Singh O. J. A. P, Shan B. P. A Review on Techniques for Computer Aided Diagnosis of Soft Markers for Detection of Down Syndrome in Ultrasound Fetal Images. Biomed Pharmacol J 2017;10(3). Available from: http://biomedpharmajournal.org/?p=15944 |
Introduction
Down Syndrome is one of the predominantchromosomal disorder caused by the presence of an extra copy of the 21st chromosome. Usually an individual possess 46 chromosomes, 23 inherited from the father and 23 from the mother. In few cases 47 chromosomes may be present in each cell instead of 46. This condition is termed as Down Syndrome.The British Doctor, John Langdon Down described the syndrome in 1866 and hence the name.1,2
The prevalence of Down syndrome is approximated to be about 1 in 1000 births worldwide.3 Earlier it has been 1 in 700.4 There are three types of Trisomy21 which are complete Trisomy 21, mosaicism and translocation. The most common one which occurs in about 95% of the cases is complete Trisomy 21 in which all the cells possess an extra copy of the chromosome 21. Inmosaicism not all the cells will have an extra chromosome and translocation happens when a whole or extra copy of chromosome occurs getting attached to another chromosome.2
No specific reasons for this disorder has been reported so far other than the maternal age.5 The likelihood of having a child with Down Syndromeincreases with increasing maternal age. Prenatal Screening of Down syndrome involves screening as well as diagnosis tests. Screening is made by noninvasive procedures like 2D/3D ultrasounds and diagnosis involves invasive methods like amniocentesis, chorionic villus sampling(CVS) and percutaneous umbilical blood sampling(PUBS). With invasive procedures there are possibilities for fetal loss and therefore they are not recommended for prenatal screening. Hence it is worthy to perform diagnosis with noninvasive procedures6 and the detection rate can be improved by combining these ultrasound findings with maternal serum markers.9,10,11,12
Two Dimensional ultrasound imaging is more popular in obstetrics and gynecology because it is noninvasive, cost effective, intuitive, convenient and safe. But the problem with ultrasound is the image quality where the original RF signal is subjected to a number of processing steps before being converted into an image as well as the multiplicative speckle noise which reduces the visibility of the image. Therefore the image presented to a physician is such a low quality image which renders it inappropriate for accurate diagnosis.7 Therefore the ultrasound images require efficient preprocessing algorithms to provide accurate results when they are to be given as inputs to clinical decision support systems.
Down Syndrome Soft Markers
A number of soft markers have been identified for chromosomal aneuploidies. The table below gives a brief picture on the literatures published on the various soft markers identified.
Table 1: Soft Markers for Down Syndrome
| Low maternal serum alpha-fetoprotein.[9]
Merkatz IR et.al[1984] |
Women suspected of having an affected fetus are subjected to serum screening tests along with normal subjects. It has been noted that those with affected fetus had maternal serum alpha-fetoprotein levels significantly lower than normal subjects. |
| Femur Length [10]
Nyberg et al[1990] |
With shortening of femur length as a marker the predictive values are 0.93 and 0.33 for high risk population and low risk population respectively. Therefore it has been concluded that this marker is less effective in screening for down syndrome. |
| Prenatal ultrasonography [11]
Nyberg DA et al [1995] |
Analysis of the effectiveness of prenatal ultrasonographic findings in screening for down syndrome when combined with the three biochemical markers–maternal serum alpha-fetoprotein, unconjugated estriol and human chorionic gonadotropin. Unusual results from these examinations reveal the necessity for an invasive screening by amniocentesis. |
| Maternal age and fetal nuchal translucency[12], Pandya PP et al [1995] | Screening for down syndrome by the combination of maternal age, crown rump length and nuchal translucency screening is proposed. |
| Free beta-human chorionicgonadotropinand pregnancy-associated plasma protein A [13]
Krantz DA et al [1996] |
Screening for down syndrome in the first trimester has been proposed which includes testing the levels of pregnancy-associated plasma protein free beta-human chorionic gonadotropin. This method is capable of producing results similar to those produced by the second trimester screening proposed in [10] |
| Serum screening for Down’s syndrome. [14]
Wald NJ et al [1996] |
Down Syndrome screening using Pregnancy Associated Plasma Protein, Serum free β-Human Chorionic Gonadotrophinand maternal age at 10 weeks produce better results than the double test mentioned in [11] and the triple test proposed in[13] |
| Combining ultrasound and biochemistry.[15]
Wald NJ et al [1997] |
The serum markers are combined with nuchal translucency measurement and the detection rate improved from 62-80 %. The 62% was obtained by combining free β-Human Chorionic Gonadotrophin and maternal age. |
| Assessment of risk of trisomy 21[16]
Snijders RJ[1998] |
Combining maternal age and nuchal translucency thicknessat a gestational age of 10 – 14 weeks achieved a detection rate of about 80% of affected pregnancies. |
| A screening programme for trisomy 21 at 10-14 weeks.[17]
Spencer[1999] |
89% accuracy with 5% false positive rate can be achieved by the combination of pregnancy associated plasma protein, maternal serum free beta human chorionic gonadotrophin, maternal age and nuchal translucency. |
| 2ndtrimester screening for DS [18]Nyberg DA et al [2001] | The following six markers are identified and evaluated: nuchal translucency thickness, shortened femur,hyperechoic bowel, shortened humerus, renal pyelectasis and echogenic intracardiac focus. |
| Nasal bone hypoplasia [19]Cicero S et al [2003] | The presence or absence of nasal bone as a useful marker for down syndrome at 15 – 22 weeks gestation has been identified. |
| Fetal tricuspid regurgitation [20]
Gustavo et al [2006] |
The association of tricuspid regurgitation with the presence of chromosomal defects is studied and the likelihood ratios in fetuses with tricuspid regurgitation for trisomy 21 and trisomy 18 are calculated. |
| DuctusVenosus in the First Trimester [21]
Wee L.K et al [2010] |
Relationship between an abnormal flow in the ductusvenosus and trisomy 21 is studied. |
| Internal application of soft markers and maternal serum markers.[22]
Ghaffari et al [2012] |
The efficiency of the first trimester screening can be improved by combining all these markers Nuchal translucency, tricuspid regurgitation, and ductusvenosusalong with the maternal serum markers thereby reducing the false positive rate from 4.8% to 3.4% |
| Fronto nasal fold thickness[23]
Gonzalez et al [2013] |
The FNF/NBL ratio is also identified as a valuable marker in the detection of Trisomy 21.It has been observed that the FNF/NBL ratio remained constant, with a mean value of 0.68, 0.84 in 95th percentile and 0.90 in the 99th percentile. |
| Fetal Pinna Measurement [24]
Rajanna et al [2016] |
A study on the Pinna length measurement and its association with chromosomal disorders has been made. There exists a linear relationship between the pinna length and the gestational age. This study is specifically made on south Indian Population and there is no remarkable difference in the values when compared. |
A few soft markers as reported by the Sandiego Perinatal Center have been discussed below:
Nuchal Translucencyis the accumulation of hypodermal fluid in the fetal neck between 11.3 and 13.6 weeks of gestation. It has been noted that babies with Down syndrome will have an increased amount of this fluid. The nuchal translucency thickness between 2.2 mm and 2.8 mm during this 10-13 weeks is considered to be normal. The risk increases with increasing translucency thickness.A visible rigid bone at the top of the nose is seen at 15 – 22 weeks of gestation. Absence of this nasal bone is also considered to be an important marker for Down Syndrome. This increases the Down Syndrome risk by a factor between 20 and 60.This can be identified only after 20 – 22 weeks of pregnancy. A linear arrangement of mitral and tricuspid valve in a baby is the symptom for some heart defect in the growing fetus. If the heart functions normal with a linear arrangement of these valves then this increases the risk for Down Syndromeby a factor of 30 to 60.
A ratio between the length of the humerus and the average humerus length is another important marker.If this is far below the expected range, the risk is increased by a factor of 6. Similar to this the femur length will also increase the risk by a factor of 2.2.Echogenic foci and a small amount of extra fluid within the baby’s kidneys are also considered to be low level markers for Down Syndrome.
The ratio of biparietal diameter/fronto nasal fold thickness to nasal bone length is also considered as another marker, where the rate of biparietal diameter to nasal bone length according to gestational week is found to be 8.1± 1.4 whereas it is 11.3+2.0 in fetuses with trisomy 21.7,16
The reference range for detecting these anomalies is being fixed by analyzing a number of measurements taken from different tertiary centres. The ethnicity also plays a major role in finalizing these reference values.25 These values may vary between different ethnic groups thereby affecting the accuracy of detection which may result in increase of false positive and false negative rates. There is no much reports specifically analyzing a group especially the South Indian population. The table below gives the normal values for few markers in reference to the South Indian population.
Table 2: Normal Values for soft Markers
| Parameters | Normal Values(mm) | Gestation(weeks) |
| Nuchal Translucency Thickness[28] | <2.12 | 10-13 |
| Nasal Bone length (Indian population)[26] | 3.3 | 16 |
| Biparietal length/Nasal Bone Diameter[10] | 9.9±1.5 | No Change |
| Biparietal Diameter/ Fronto Nasal fold Thickness [27] | 8.1±1.4 | No Change |
| Pinna Length(South Indian Population)[24] | 8.1-29.5 | 15 – 28 |
Automated Diagnosis
Accurate detection of the above markers require skilled sonographers, obstetricians and fetal medicine professionals, since the ultrasound markers can be easily confused with the underlying structures because of the speckle noise. Researchers have been working on automated diagnosis of these soft markers so that observer dependence will be minimized and the detection rate could be improved. If these parameters could be estimated from a B mode image by computer assisted techniques the detection accuracy will be higher and the number of false positive rates or false negative rates could be reduced.
Yenhui Deng et al has proposed an ordered structural model for the automated detection of the nuchal translucency region.28 Anzalone et al has proposed a completely automated system for Nuchal Translucency measurement which operates on the image sequence what we get as output from the ultrasound machine.29 S. Nirmala and V. Palanisamy have utilized the mean shift analysis and canny operators for segmentation of the nuchal translucency region. The exact thickness of the nuchal translucency region has been estimated using blob analysis. Their results say that the fetus should have a nuchal translucency thickness of 1.85±0.2 mm in the 14th week of gestation.30 R.Sonia and V. Shanthi[31]propose a computerized method to measure nuchal translucency thickness. Preprocessing is done using Lee filter. Region of Interest is manually extracted. Segmentation is done using morphological operations and otsuthresholding. The Average height and standard deviation of Nuchal Translucency thickness for normal fetus is 1.99 ± 0.62mm and for abnormal fetus is 4.10 ± 9.00 mm respectively. For area it is found to be 37.84 ± 20.28 mm and 126.44 ± 41.80 mm.
Lai K W et al [32] have trained the Artificial neural network to locate the region of interest that contains the Nuchal Translucency. The accuracy achieved is 93.33 percentage. The boundary region of the Nuchal Translucency layer is identified using instinctive computerized algorithm. Once the boundary region is located then the optimum thickness of the region is determined. Intensity continuity and edge strength are the local parameters used as biased terms for thickness calculation.Moratalla J et al[33] a segmentation algorithm based on minimizing a cost function is proposed. This method is semi-automatic where the region of interest is selected manually. The boundary selection is based on minimization of a cost function. The dynamic programming technique is employed for optimization.
Park JH et al[34] has proposed a fully automatic approach for computing Nuchal Translucency (NT) measurement in ultrasound scans of the mid-sagittal plane of a fetal head. The algorithm finds fetal head using discriminative learning-based detectors. The NT region is estimated from the statistical relationship between the fetal head and the NT region. The boundaries of the nuchal translucency region are determined by Dijkstra’s shortest path applied on the edge-enhanced image. Finally, these two region edges are used to define foreground and background seeds for accurate graph cut segmentation. The NT measurement is computed from the segmented region.
The significance of neural networks in feature extraction of ultrasound fetal images has been analysed by Neocleous et al.35 A number of artificial neural network schemes, support vector machines and K- nearest neighbour models are developed and tested which revealed that ANN’s outperformed the other networks with 0% false negative rate for T21 and identified 96.1% of euploidies with 3.9% false positive rate. Anjit&Rishidas have utilized the backpropagation artificial neural network for the detection of presence or absence of nasal bone. The features are extracted in spatial domain and transform domain using discrete cosine transforms and wavelet transforms and they are provided an inputs to train a BPN. The results obtained shows higher degree of accuracy in transform domain than spatial domain.36
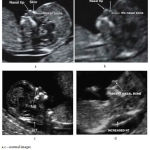 |
Figure 1: Normal and affected fetal images |
Lai KWet al37 has worked out a mathematical model which combines three maternal serum markers using trivariate log normal distribution and automatically calculates the likelihood of having a fetus with down syndrome have been introduced. A semiautomatic measurement of fetal nuchal translucency has been proposed with the development of a software package by Bernardino et al.38 An enhancing diffusion filter has been applied to enhance the border and reduce noise in.39 The NT is detected by minimization of a cost function that combines intensity, edge strength and continuity using dynamic programming. Fully automatic measurement of biparietal diameter (BPD), abdominal circumference, head circumference (HC), humerus length, femur length and crown rump length has been proposed in.40 The nuchal translucency thickness varies with gestational age as well as crown rump length. These discrepancies should also be taken into consideration while obtaining normative values for anomaly detection.41 presents a reference value for nuchal translucency thickness with respect to gestational age and crown rump length.
Automated detection of nasal bone has not been much reported so far. Lai Khin Wee et al have presented a method to recognize and detect the fetal nasal bone based on 2D ultrasound images using cross correlation techniques. The threshold is set 0.35 for classifying the presence or absence of nasal bone. The accuracy achieved by this technique is 96.26.42
Table 3: Performance Measures of various computer aided techniques
| S.No | Author | Method | No. of subjects | Accuracy(%) | Sensitivity | Specificity | Detection Rate |
| 1 | Andreas C Neocleous | Artificial Neural Networks | 129 | 96 | |||
| 2 | Lai Khin Wee | Cross correlation techniques | 107 | 96.26 | 97 | 85.77 | |
| 3 | A. Khashman | Neural Networks | 26 | 92 | |||
| 4 | C.K. Neocleous | Neural Networks | 71 | 80.3 | 98.6 | 78.9 | |
| 5 | Anjit T. A | Neural Networks | 50(with nasal bone | 86 | |||
| 50(without nasal bone) | 88 | ||||||
| 6 | EkoSupriyanto | Neural Networks | 100 | 93.3 |
An important step in feature extraction from ultrasound images is extraction of the region of interest. A number of approaches have been proposed in literatures for efficient segmentation of the underlying region of interest. Table below gives a review of few algorithms proposed for segmentation of ultrasound fetal images.
Table 4: Segmentation Algorithms for DS markers
| KalpathiR.Subramanianet al [43] | Segmentation by region growing as well as split and merge algorithms with slight variant applied are explored and the results show that upto 50 images can be segmented in 5-6 minutes. |
| ShaziaAnjumet al [44] | Multilevel thresholding and multilevel thresholding with smoothing are compared. The experimental results show that the later one performs better. |
| VibhakarShrimaliet al [45] | The femur is separated from the background using morphological operators to obtain a single pixel wide skeleton of the femur. The observed results are consistent and in good agreement with conventional manual method of measurement. |
| Lalit Guptaet al [46] | The low quality of US images are taken into consideration and therefore the intensity variations of different tissues along with their shape priors are utilized in the optimization function. |
| Sonia Dahdouhet al [47] | B-spline two dimensional wavelet transform is utilized. Feature vectors are generated for each pixel with the following parameters gray level, moments and texture. These parameters are given as input for fuzzy C means clustering. |
| NourhanZayedet al [48] | The Nuchal translucency region is extracted by region growing segmentation technique based on threshold boundary computation. |
| SiqingNieet al [49] | The presence of fetal head in ultrasound images is detected by training a network, the deep belief network and the position as well as size is measured by the use of modified circle detection method. |
| Angee Paola et al [50] | A hierarchial segmentation technique is adopted, where initially the fetal nose is identified and then the three lines in the nasal region. The presence of nose is recognized using the combination of regional maxima and high eccentricity detection algorithms. The nasal lines using morphological operators and k means clustering algorithms. |
Conclusion
A review on the various soft markers identified for the detection of down syndrome from the first and second trimester ultrasound fetal images has been made. The normal and abnormal values for these markers has been studied. It could be noted from literatures that the diagnostic accuracy can be improved by the combinational analysis of two or more soft markers rather than relying on a single marker alone. A brief survey on few segmentation schemes on ultrasound images has been made.
References
- Regezi S. Oral pathology. Clinical pathologic correlations, 1st ed. Philadelphia: Saunders. 1989:450-51.
- kcdsg.org/files/content/About%20Down%20Syndrome.pdf
- http://www.who.int/genomics/public/geneticdiseases/en/index1.html
- cdc.gov/ncbddd/birthdefects/downsyndrome/data.html
- http://www. medicalnewstoday.com/articles/145554. php
- Leshin L. Prenatal Screening for Down Syndrome. 2007
- Wachinger C., Klein T and Navab N. The 2D analytic signal for envelope detection and feature extraction on ultrasound images elsevier. Medical Image Analysis. 2012;16:1073–1084.
CrossRef - ResulArsoy., NidaErgin., Yayla M., GökhanGöynümer. The Ratio of Biparietal Diameter to Nasal Bone Length. Perinatal Journal. 2010;18(3).
- Merkatz I. R.,Nitowsky H. M., Macri J. N., Johnson W. E. An association between low maternal serum alpha-fetoprotein and fetal chromosomal abnormalities.American Journal of Obstetrics and Gynecology. 19841;148(7):886-94.
- Nyberg D. A et al. Femur length shortening in the detection of Down syndrome: is prenatal screening feasible. American Journal of Obstetrics and Gynecology. 1990;162(5):1247-52.
CrossRef - Nyberg D. A., Luthy D. .A, Cheng E. Y et al. Role of prenatal ultrasonography in women with positive screen for Down syndrome on the basis of maternal serum markers. American Journal of obstetrics and Gynecology. 1995;173:1030-5.
CrossRef - Pandya P. P.,Snijders R. J., Johnson S .P., De Brizot M. L., Nicolaides K. H. Screening for fetal trisomies by maternal age and fetal nuchal translucency thickness at 10 to 14 weeks of gestation.British Journal of Obstetrics and Gynaecology. 1995 Dec;102(12):957-62.
CrossRef - Krantz D. A.,Larsen J.W., Buchanan P. D., Macri J. N. First-trimester Down syndrome screening: free beta-human chorionic gonadotropin and pregnancy-associated plasma protein A. American Journal of obstetrics and Gynecology. 1996;174(2):612-6.
CrossRef - Wald N. J., George L., Smith D et al. Serum screening for Down’s syndrome between 8 and 14 weeks of pregnancy. British Journal of Obstetrics and Gynaecology. 1996;103:407-11
CrossRef - Wald N. J., Hackshaw A. K. Combining ultrasound and biochemistry in first-trimester screening for Down’s syndrome. Prenatal Diagnosis. 1997;17:821-9. 26.
- Snijders R. J.,Noble P., Sebire N., Souka A., Nicolaides K. H. UK multicentre project on assessment of risk of trisomy 21 by maternal age and fetal nuchal-translucency thickness at 10-14 weeks of gestation. 1998;352(9125):343-6.
- Spencer K., Souter V., Tul N et al. A screening programme for trisomy 21 at 10-14 weeks using fetal nuchal translucency, maternal serum free â-human chorionic gonadotrophin and pregnancy associated plasma protein-A.Ultrasound in Obstetrics and Gynecology. 1999;13:231-7. 27.
- Nyberg D. A.,Souter V.L., El-Bastawissi A., Young S., Luthhardt F., Luthy D. A., “Isolated sonographic markers for detection of fetal Down syndrome in the second trimester of pregnancy.Journal of Ultrasound in Medicine. 2001;20(10):1053-63.
CrossRef - Cicero S.,Sonek J. D., McKenna D. S., Croom C. S., Johnson L., Nicolaides K. H. Nasal bone hypoplasia in trisomy 21 at 15-22 weeks gestation.Ultrasound in Obstetrics and Gynecology. 2003;21(1):15-8.
CrossRef - Falcon S., Faiola I., Allan l. H and Nicolaides K. H. Fetal tricuspid regurgitation at the 11 + 0 to 13 + 6-week scan: association with chromosomal defects and reproducibility of the method. Ultrasound in Obstetrics and Gynecology. 2006;27:609–612.
CrossRef - Maiz N., Nicolaides K. H., Sebastián S. Ductus Venosus in the First Trimester: Contribution to Screening of Chromosomal, Cardiac Defects and Monochorionic Twin Complications. Mini-Review Fetal Diagnosis and Therapy. 2010;28:65–71.
CrossRef - Ghaffari R., et al. First-trimester screening for chromosomal abnormalities by integrated application of nuchal translucency nasal bone tricuspid regurgitation and ductusvenosus flow combined with maternal serum free β-hCG and PAPP-A: a 5-year prospective study. Ultrasound in Obstetrics and Gynecology. 2012;39: 528–534.
CrossRef - Gonzalez R.,Aedo S., Dezerega V., Sepulveda W. Frontonasal fold thickness-to-nasal bone length ratio as a prenatal sonographic marker for trisomy 21 in a low-risk population. Journal of Ultrasound in medicine. 2013;32(5):795-800.
CrossRef - Ambresh R.,Sharath M. Antenatal Ultrasound Fetal Pinna Measurement and Evolving Nomogram in South Indian Population in Normal Pregnancies. International Journal of Anatomy, Radiology and Surgery. 2016;5(4).
- Alphonse J.,Cox J.,Clarke J.,Schluter P and McLennan A. The Effect of Ethnicity on 2D and 3D Frontomaxillary Facial Angle Measurement in the First Trimester, Hindawi Publishing Corporation. Obstetrics and Gynecology International. 2013;5. Article ID 847293.
- NarayaniBandeppa H., RadhakrishnanPrathima. Mid-second Trimester Measurement of Nasal Bone Length in the Indian Population.The Journal of Obstetrics and Gynecology of India. 2013.
- BromleyB., Lieberman E.,Shipp T. D.,Benacerraf B. R. Fetal Nose Bone Length A Marker for Down Syndrome in the Second Trimester.Journal of Ultrasound and Medicine. 2002;21:1387–1394.
CrossRef - Deng Y., Wang Y., Chen P. Automated detection of fetal nuchal translucency based on hierarchical structural model”, IEEE International Symposium on Computer-Based Medical Systems (CBMS); Perth, WA. 2010;78–84.
CrossRef - Anzalone et al., A system for the automatic measurement of the nuchal translucency thickness from ultrasound video stream of the foetus. Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems; 2013;(s):239 –244.
CrossRef - Nirmala S., Palanisamy V. Measurement of nuchal translucency thickness for detection of chromosomal abnormalities using first trimester ultrasound fetal images. International Journal of Computer Science and Information Security. 2009;6(3):101–6.
- Shanthi V. S. Early Detection of Down Syndrome Marker by Measuring Fetal Nuchal Translucency Thickness from Ultrasound Images during First Trimester.Indian Journal of Science and Technology. 2016;9(21):1-6.
- Lai K. W., Min T. Y., Arooj A.,Supriyanto E. Nuchal translucency marker detection based on artificial neural network and measurement via bidirectional iteration forward propagation. WSEAS Transactions on Information Science and Applications. 2010;7(8):1025–36.
- Moratalla J et al. Semi-automated system for measurement of nuchal translucency thickness. Ultrasound in Obstetrics and Gynecology. 2010;36(4):412–6.
CrossRef - Park J. H et al., Automatic nuchal translucency measurement from ultrasonography. Medical Image Computing and Computer-Assisted Intervention- Springer-Verlag: Berlin Heidelberg. 2013;16(3):243–50.
CrossRef - Neocleous K., Nicolaides K. H.,Neokleous K.C and Schizas C. N. Artificial neural networks for non-invasive chromosomal abnormality screening of fetuses. IEEE. 2010
CrossRef - Anjit T. A., Rishidas S. Identification of Nasal Bone for the Early Detection of Down Syndrome using Back Propagation Neural Network” IEEE. 2011.
- Lai K. W., Supriyanto E. Automated risk calculation for trisomy 21 based on maternal serum markers using trivariate lognormal distribution” Proceedings of the 12th WSEAS International Conference on Automatic control, modelling & simulation. 2010.
- Bernardino F., Cardoso R.,Montenegro N.,Bernardes J and marques de sa J “Semiautomatedultrasonographic measurement of fetal Nuchal translucency using a computer software tool. Ultrasound in Medicine and Biology. 1998;24(1):51-54.
CrossRef - Yu-Bu L., Min-Jeong K., Myoung-Hee K., Robust border enhancement and detection for measurement of fetal nuchal translucency in ultrasound images, Medical and Biological Engineering and Computing. 2007;45:1143–1152.
CrossRef - Carneiro G., Georgescu B.,Good S., Comaniciu D. Detection of Fetal Anatomies from Ultrasound Images using a Constrained Probabilistic Boosting Tree. IEEE. 2008:1-13.
- Mahale N., Kumar A., Rayapureddi C. V. M., Mahale A. Variaton of nuchal translucency with increasing crown rump length and gestational age in normal singleton pregnancies. IOSR Journal of Dental and Medical Sciences. 2013;6(3):16-19.
CrossRef - Wee L. K., Min T. Y., Arooj A., Supriyanto E. Computerized Automatic Nasal Bone Detection based on Ultrasound Fetal Images Using Cross Correlation Techniques. WSEAS Transactions on Information Science and Applications. 2010;7(8):1068–77.
- Dina M. S.,TaghiMostafavi L. M. Interactive Segmentation and Analysis of Fetal Ultrasound Images, 8th EG Workshop on ViSC. Boulogne sur. Mer. 1997;28-30.
- Anjum S., Pavithra G., Manjunath Dr. T. C. Efficient Segmentation of the Foetal Ultrasound Image Using Smoothing Algorithm. International Journal of Innovative Research in Computer and Communication Engineering. 2015;3:4.
- VibhakarShrimali R. S. A and Kumar V. Improved Segmentation of Ultrasound Images for Fetal Biometry Using Morphological Operators” 31st Annual International Conference of the IEEE EMBS Minneapolis, Minnesota, USA. 2009;2-6.
- Gupta L., Sisodia R. S., Pallavi V., Firtion C and Ramachandran G. Segmentation of 2D Fetal Ultrasound Images by Exploiting Context Information using Conditional Random Fields” 33rd Annual International Conference of the IEEE EMBS Boston, Massachusetts USA, August 30 – September 3. 2011.
- Dahdouh S., Serrurier A., Grang´ey G., Angelini D. E., Bloch I. Segmentation of Fetal Envelope from 3d Ultrasound Images based on Pixel Intensity Statistical Distribution and Shape Priors. IEEE 10th International Symposium on Biomedical Imaging. 2013.
- Zayed N., Badawi A., Elsayad A. Wavelet segmentation for fetal ultrasound Images” From Nano to Macro, San Francisco, CA, USA, April 7-11. 2013,
- Nie S., Yu J., Chen P., Zhang J., Wang Y. A Novel method with a deep network and directional edges for automatic detection of a fetal head” IEEE. 2015.
- Paola A et al. Segmentation and nasal line counting in ultrasound fetal images at the 11-13+6 weeks of gestation. Ingeniería. 2015;20(1).







